
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jitka Richtová | + 2753 word(s) | 2753 | 2021-06-25 08:01:26 | | | |
| 2 | Rita Xu | -30 word(s) | 2723 | 2021-07-08 09:02:40 | | |
Video Upload Options
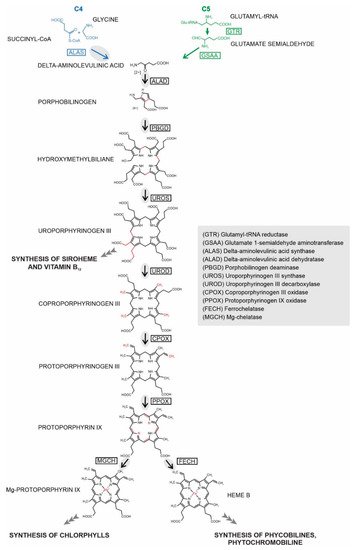
Heme biosynthesis is essential for almost all living organisms. Despite its conserved function, the pathway’s enzymes can be located in a remarkable diversity of cellular compartments in different organisms. This location does not always reflect their evolutionary origins, as might be expected from the history of their acquisition through endosymbiosis. Instead, the final subcellular localization of the enzyme reflects multiple factors, including evolutionary origin, demand for the product, availability of the substrate, and mechanism of pathway regulation. Chromera velia is a coral-associated alga bearing complex rhodophyte-derived plastid with a peculiar tetrapyrrole pathway. It synthesizes ALA using heterotrophic C4 path (same as apicomplexan parasites), which additionally supplies chlorophyll for photosystems. Using a combination of bioinformatics and experimental approaches, we investigated localizations of heme pathway enzymes in C. velia. Our data show that the pathway very likely starts in the mitochondrion, with the remaining enzymes located in the plastid. We demonstrate that the proteins are targeted to various cellular compartments by stringent translocon mechanisms that are not universal even for evolutionarily related organisms.
1. Introduction
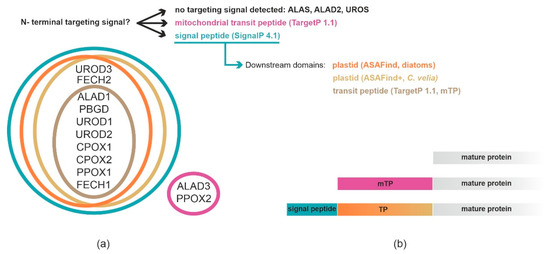
2. Prediction of Localization of Heme Synthesis Enzymes in C. velia
3. Analyses of C. velia Heme Pathway Enzymes N-termini Sequence
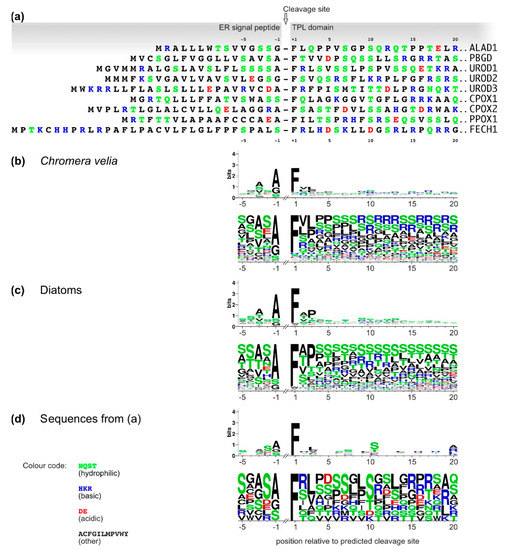
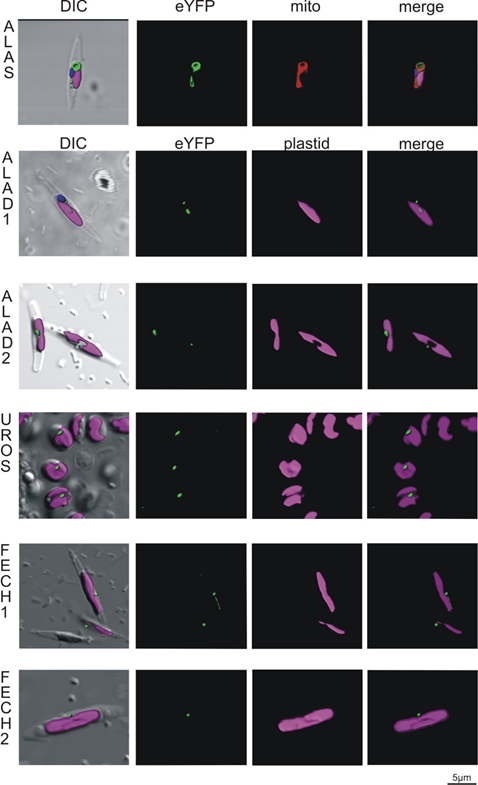
Figure 4. Heterologous expression of Phaeodactylum tricornutum with genes from Chromera velia heme pathway enzymes. Selected enzymes were tagged on their C-terminus by eYFP (green), magenta indicates plastid autofluorescence, MitoTracker® Orange CM-H2TMRos (ALAS, red) indicates mitochondrion. The Green eYFP signal of C. velia ALA synthase colocalizes with the red signal of P. tricornutum mitochondrion (row ALAS). Typical “blob-like” structures are found in heterologous expression of ALA dehydratases (ALAD1, ALAD2), uroporphyrinogen synthase (UROS), and both ferrochelatases (FECH1, FECH2).
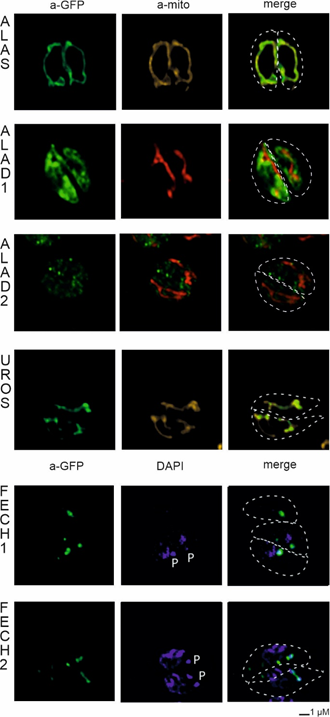
Figure 5. Heterologous expression of Toxoplasma gondii with genes from Chromera velia heme pathway enzymes. Immunofluorescence assays of transfected T. gondii, anti-GFP antibody were used to detect eYFP tagged C. velia enzymes. Anti-GFP (green) colocalized with mitochondrial anti-TgMys (a-mito, red and yellow) signal in case of ALAS and UROS. ALAD1 and ALAD2 signal were detected in the cytosol. FECH1 and FECH2 signal was found to overlap with DAPI (blue) signal at the area of parasite apicoplast. The apicoplast is denoted by “P.” Dashed line indicates T. gondii cell border.
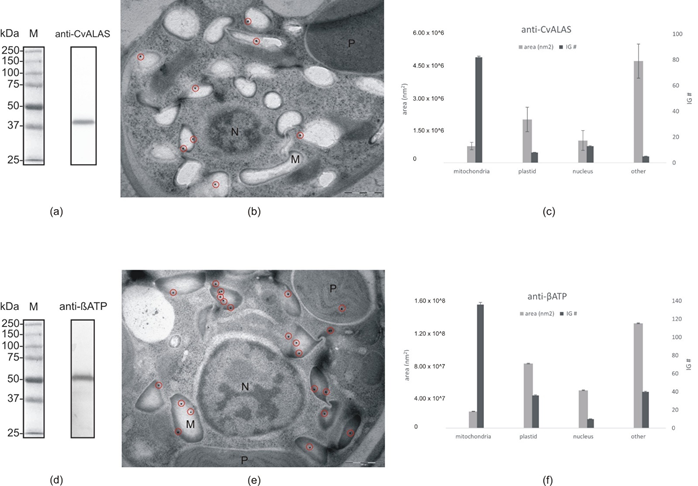
Figure 7. Immunogold labeling: (a) Western blot with anti-CvALAS on total protein extract from C. velia (b) Micrograph of C. velia ultrathin section after immunogold labeling with specific anti-CvALAS as a primary antibody. The majority of gold particles (encircled) were detected in the mitochondria. (c) Distribution of secondary IG particles (detecting anti-CvALAS) among cell compartments counted from all 35 micrographs. (d) Western blot with anti-βATP on total protein extract from C. velia (e) Micrograph of C. velia ultrathin section after immunogold labeling with specific anti-βATP as a primary antibody. The majority of gold particles (encircled) were detected in C. velia mitochondria. (f) Distribution of secondary IG particles (detecting anti-βATP) among cell compartments counted from all 35 micrographs. N = nucleus, M = mitochondria, P = plastid.
|
Enzyme |
Accession (CryptoDB) |
Evolutionary origin |
Targeting prediction |
Localization |
Localization |
Conclusion |
|
|
|
ALAS |
Cvel_28814.t1 |
Alphaproteobacteria |
No targeting signal identified |
Mitochondria |
Mitochondria |
Mitochondria |
||
|
ALAD1 |
Cvel_108.t1 |
Cyanobacteria |
Plastid |
Cytosol |
PPC/ER |
Plastid |
||
|
ALAD2 |
Cvel_13826.t1 |
Primary alga |
No targeting signal identified |
Cytosol |
PPC/ER |
Uncertain location |
||
|
ALAD3 |
Cvel_36189.t1 |
Proteobacterial |
Mitochondria |
Not tested |
Not tested |
Uncertain location |
||
|
PBGD |
Cvel_26028.t1 |
Alphaproteobacteria |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
UROS |
Cvel_15018.t1 |
Uncertain origin in primary alga |
No targeting signal identified |
Mitochondria |
PPC/ER |
Uncertain location |
||
|
UROD1 |
Cvel_14720.t1 |
Cyanobacteria |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
UROD2 |
Cvel_5098.t1 |
Endosymbiont nucleus |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
UROD3 |
Cvel_31936.t1 |
Secondary host nucleus |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
CPOX1 |
Cvel_21486.t1 |
Secondary host nucleus |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
CPOX2 |
Cvel_2641.t1 |
Uncertain origin in primary alga |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
PPOX1 |
Cvel_13840.t1 |
Cyanobacteria |
Plastid |
Not tested |
Not tested |
Plastid |
||
|
PPOX2 |
Cvel_18037.t1 |
Eukaryotic origin |
Mitochondria |
Not tested |
Not tested |
Uncertain location |
||
|
FECH1 |
Cvel_18167.t1 |
Cyanobacteria |
Plastid |
Apicoplast |
PPC/ER |
Plastid |
||
|
FECH2 |
Cvel_26873.t1 |
Alphaproteobacteria |
Signal peptide positive |
Apicoplast |
PPC/ER |
Uncertain location |
||
Table showing results described in this manuscript. Enzymes are listed according to their order during the synthesis of heme. The evolutionary origins of each of the enzymes are based on phylogenetic analyses from the work of [11][16]. The last column of the table contains our hypothetical conclusions about the C. velia enzyme localization based on our findings.
While the tetrapyrrole pathway starts with the ALAS in the mitochondrion in chromerids, the remaining steps likely occur in the plastid. This model is further supported by the phylogenetic relationships among the individual enzymes of the pathway [11]. We summarized our findings in Table 1. The heterologous expression of C. velia ALAD1 and ALAD2 gave the same inconsistent results, placing the protein in the cytosol of T. gondii and PPC/ER in P. tricornutum. Despite that, we assume that ALAD1 is more likely a plastid-targeted protein because our experimental results in P. tricornutum showed that the construct was transferred at least through the two outermost membranes of the diatom plastid. This, together with the combination of its cyanobacterial evolutionary origin, leads us to the conclusion that plastid localization is more plausible. The same cogitation was applied for FECH1 where the corresponding enzyme is also of cyanobacterial origin, and when heterologously expressed, it localized to PPC/ER of P. tricornutum and also to the apicoplast of T. gondii. We decided to conclude with an “uncertain localization” statement for ALAD3 and PPOX2 due to the absence of the experimental evidence and predictable ER signal peptides (see Supplementary Table S2 for details), and their proteobacterial and eukaryotic origin, respectively. Both enzymes possess predicted mitochondrial transit peptides; however, particularly in PPOX, which makes a complex with FECH, its placement in the mitochondrion without FECH is unlikely. The localization of ALAD3 in the mitochondrion and a formation of porphobilinogen in this organelle would require additional transport of porphobilinogen to the plastid over the four membraned envelopes. The remaining enzymes (PBGD, UROD1, UROD2, UROD3, CPOX1, CPOX2, and PPOX1) were concluded as “plastid” localized due to the congruency of the prediction result. However, spatial separation of the beginning and the end of the pathway is unprecedented, and it would require regulatory mechanisms that are not yet known. Therefore, we cannot rule out the possibility of recent reassignments of intracellular locations or dual targeting of the enzymes. Our work on localization of C. velia heme pathway enzymes shows that the subcellular localization of biosynthetic pathway within any organism is a concert of multiple factors rather than a solo for one major element.
References
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346.
- Koreny, L.; Sobotka, R.; Kovarova, J.; Gnipova, A.; Flegontov, P.; Horvath, A.; Obornik, M.; Ayala, F.J.; Lukes, J. Aerobic kinetoplastid flagellate Phytomonas does not require heme for viability. Proc. Natl. Acad. Sci. USA 2012, 109, 3808–3813.
- Jordan, P.M. Highlights in haem biosynthesis. Curr. Opin. Struct. Biol. 1994, 4, 902–911.
- Oborník, M.; Green, B.R. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 2005, 22, 2343–2353.
- Kořený, L.; Oborník, M.; Lukeš, J. Make it, take it, or leave it: Heme metabolism of parasites. PLoS Pathog. 2013, 9, e1003088.
- Masuda, T.; Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008, 7, 1131–1149.
- Czarnecki, O.; Grimm, B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 2012, 63, 1675–1687.
- Ikushiro, H.; Nagami, A.; Takai, T.; Sawai, T.; Shimeno, Y.; Hori, H.; Miyahara, I.; Kamiya, N.; Yano, T. heme-dependent inactivation of 5-aminolevulinate synthase from Caulobacter crescentus. Sci. Rep. 2018, 8, 1–13.
- Wißbrock, A.; George, A.A.P.; Brewitz, H.H.; Kühl, T.; Imhof, D. The molecular basis of transient heme-protein interactions: Analysis, concept and implementation. Biosci. Rep. 2019, 39.
- Kloehn, J.; Harding, C.R.; Soldati-Favre, D. Supply and demand—Heme synthesis, salvage and utilization by Apicomplexa. FEBS J. 2021, 288, 382–404.
- Kořený, L.; Sobotka, R.; Janouškovec, J.; Keeling, P.J.; Oborník, M. Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell 2011, 23, 3454–3462.
- Hamza, I. Intracellular trafficking of porphyrins. ACS Chem. Biol. 2006, 1, 627–629.
- Swenson, S.A.; Moore, C.M.; Marcero, J.R.; Medlock, A.E.; Reddi, A.R.; Khalimonchuk, O. From synthesis to utilization: The ins and outs of mitochondrial heme. Cells 2020, 9, 579.
- Mochizuki, N.; Tanaka, R.; Grimm, B.; Masuda, T.; Moulin, M.; Smith, A.G.; Tanaka, A.; Terry, M.J. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010, 15, 488–498.
- Kořený, L.; Oborník, M. Sequence evidence for the presence of two tetrapyrrole pathways in Euglena gracilis. Genome Biol. Evol. 2011, 3, 359–364.
- Cihlář, J.; Füssy, Z.; Horak, A.; Oborník, M. Evolution of the tetrapyrrole biosynthetic pathway in secondary algae: Conservation, redundancy and replacement. PLoS ONE 2016, 11, e0166338.
- Votýpka, J.; Modrý, D.; Oborník, M.; Šlapeta, J.; Lukeš, J. Apicomplexa. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 567–624.
- Ralph, S.A.; Van Dooren, G.G.; Waller, R.; Crawford, M.J.; Fraunholz, M.; Foth, B.J.; Tonkin, C.J.; Roos, D.; McFadden, G.I. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004, 2, 203–216.
- Seeber, F.; Limenitakis, J.; Soldati-Favre, D. Apicomplexan mitochondrial metabolism: A story of gains, losses and retentions. Trends Parasitol. 2008, 24, 468–478.
- Van Dooren, G.G.; Kennedy, A.T.; McFadden, G.I. The use and abuse of heme in apicomplexan parasites. Antioxid. Redox Signal. 2012, 17, 634–656.
- Tjhin, E.T.; Hayward, J.A.; McFadden, G.I.; van Dooren, G.G. Characterization of the apicoplast-localized enzyme TgUroD in Toxoplasma gondii reveals a key role of the apicoplast in heme biosynthesis. J. Biol. Chem. 2020, 295, 1539–1550.
- Dalbey, R.; von Heijne, G. Protein Targeting, Transport, and Translocation, 1st ed.; Academic Press: Cambridge, MA, USA, 2002; 336p.
- Kroth, P.G. Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int. Rev. Cytol. 2002, 221, 191–255.
- Gould, S.B.; Waller, R.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517.
- Bolte, K.; Bullmann, L.; Hempel, F.; Bozarth, A.; Zauner, S.; Maier, U.G. Protein targeting into secondary plastids. J. Eukaryot. Microbiol. 2009, 56, 9–15.
- Maier, U.G.; Zauner, S.; Hempel, F. Protein import into complex plastids: Cellular organization of higher complexity. Eur. J. Cell Biol. 2015, 94, 340–348.
- Cavalier-Smith, T. Kingdom Chromista and its eight phyla: A new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 2018, 255, 297–357.
- Moore, R.B.; Oborník, M.; Janouškovec, J.; Chrudimský, T.; Vancová, M.; Green, D.H.; Wright, S.W.; Davies, N.W.; Bolch, C.J.S.; Heimann, K.; et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 2008, 451, 959–963.
- Oborník, M.; Modrý, D.; Lukeš, M.; Černotíková-Stříbrná, E.; Cihlář, J.; Tesařová, M.; Kotabová, E.; Vancová, M.; Prasil, O.; Lukeš, J. Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist 2012, 163, 306–323.
- Janouškovec, J.; Tikhonenkov, D.; Burki, F.; Howe, A.T.; Kolísko, M.; Mylnikov, A.P.; Keeling, P.J. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. USA 2015, 112, 10200–10207.
- Oborník, M.; Kručinská, J.; Esson, H. Life cycles of chromerids resemble those of colpodellids and apicomplexan parasites. Perspect. Phycol. 2016, 3, 21–27.
- Füssy, Z.; Masařová, P.; Kručinská, J.; Esson, H.J.; Oborník, M. Budding of the alveolate alga Vitrella brassicaformis resembles sexual and asexual processes in apicomplexan parasites. Protist 2017, 168, 80–91.
- Oborník, M. Endosymbiotic evolution of algae, secondary heterotrophy and parasitism. Biomolecules 2019, 9, 266.
- Oborník, M. Photoparasitism as an intermediate state in the evolution of apicomplexan parasites. Trends Parasitol. 2020, 36, 727–734.
- Kilian, O.; Kroth, P.G. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005, 41, 175–183.
- Patron, N.J.; Waller, R. Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays 2007, 29, 1048–1058.
- Apt, K.E.; Grossman, A.R.; Kroth-Pancic, P.G. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. MGG 1996, 252, 572–579.
- Striepen, B.; He, C.Y.; Matrajt, M.; Soldati-Favre, D.; Roos, D. Expression, selection, and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol. 1998, 92, 325–338.
- Poulsen, N.; Chesley, P.M.; Kröger, N. MOLECULAR genetic manipulation of the diatom Thalassiosira pseudonana (bacillariophyceae). J. Phycol. 2006, 42, 1059–1065.
- Striepen, B.; Soldati, D. Genetic manipulation of Toxoplasma gondii. In Toxoplasma Gondii; Academic Press: Cambridge, MA, USA, 2007; pp. 391–418.
- Zhang, C.; Hu, H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genom. 2014, 16, 63–66.
- McFadden, G.I. Plastids and protein targeting. J. Eukaryot. Microbiol. 1999, 46, 339–346.
- Roos, D.S.; Crawford, M.J.; Donald, R.G.; Kissinger, J.; Klimczak, L.J.; Striepen, B. Origin, targeting, and function of the apicomplexan plastid. Curr. Opin. Microbiol. 1999, 2, 426–432.
- DeRocher, A.; Hagen, C.B.; Froehlich, J.E.; Feagin, J.E.; Parsons, M. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J. Cell Sci. 2000, 113, 3969–3977.
- Waller, R.F.; Reed, M.B.; Cowman, A.F.; McFadden, G.I. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000, 19, 1794–1802.
- Apt, K.E.; Zaslavkaia, L.; Lippmeier, J.C.; Lang, M.; Kilian, O.; Wetherbee, R.; Grossman, A.R.; Kroth, P.G. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 2002, 115, 4061–4069.
- Sheiner, L.; Demerly, J.L.; Poulsen, N.; Beatty, W.L.; Lucas, O.; Behnke, M.; White, M.W.; Striepen, B. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLOS Pathog. 2011, 7, e1002392.
- Huesgen, P.F.; Alami, M.; Lange, P.F.; Foster, L.J.; Schröder, W.P.; Overall, C.M.; Green, B.R. Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS ONE 2013, 8, e74483.
- Füssy, Z.; Oborník, M. Chromerids and their plastids. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 84, pp. 187–218.
- Oborník, M.; Lukeš, J. Cell biology of chromerids. In International Review of Cell and Molecular Biology; Academic Press: Cambridge, MA, USA, 2013; Volume 306, pp. 333–369.
- Nielsen, H. Predicting secretory proteins with SignalP. In Protein Function Prediction; Kihara, D., Ed.; Humana Press: New York, NY, USA, 2017; pp. 59–73.
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H.A. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971.
- Gruber, A.; Rocap, G.; Kroth, P.G.; Armbrust, E.V.; Mock, T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015, 81, 519–528.
- Füssy, Z.; Faitová, T.; Oborník, M. subcellular compartments interplay for carbon and nitrogen allocation in Chromera velia and Vitrella brassicaformis. Genome Biol. Evol. 2019, 11, 1765–1779.
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126.
- Gruber, A.; McKay, C.; Kroth, P.G.; Armbrust, E.V.; Mock, T. Comparison of different versions of SignalP and TargetP for diatom plastid protein predictions with ASAFind. Matters 2020, 81, 519–528.




