Due to their repetitive nature, high similarity, and large quantities in the genome, TEs present a significant substrate for non-allelic gene conversion. Gene conversion is a process whereby the genetic material of a donor sequence unidirectionally replaces that of a homologous acceptor sequence via recombination after a double-strand DNA break. Thus, gene conversion can proliferate mutations among TEs independently of the activity of the master-copy, leading to TE homogenization, a phenomenon known as concerted evolution
[5]. Earlier retrotransposon studies reported a few cases of gene conversion between TE copies. For example, Kass and colleagues
[6] described a case of gene conversion that changed a younger human
Alu SINE to an older element. Roy et al.
[7] suggested that gene conversion is responsible for ~10–20% of the variation in the young
AluYa5 subfamily. A whole-genome gene conversion analysis among
Alus in humans
[8] focused on non-diagnostic mutations in
Alu sequences revealed significant levels of gene conversion, especially among neighboring
Alus. The authors found that gene conversion acts on
Alus within a range of about 10 kb, inversely proportional to their distance from one another. Most studies of gene conversion between TEs focused on
Alu SINEs in primates. However, similar effects were also reported for LTRs in other mammals
[9][10] and in plants
[11].
The extent of sequence similarity between donor and acceptor loci positively influences the frequency of gene conversion, and reaches an optimum at 89%–100%
[17][18]. Therefore, a substantial number of gene conversion events involves young TEs of the same subfamily.
Alu elements are the most abundant TEs in primate genomes and have served as a model group for TE-based gene conversion studies e.g.,
[7][8].
Alus evolved from 7 SL RNA around 65 million years ago in the ancestral lineage of primates and consist of dimeric sequences of about 300 nt (merged 5′- and 3′-monomers
[19]). They diverged into three subfamilies/types—the oldest
AluJ, the
AluS, and the youngest
AluY. More than a million
Alu copies are distributed across the human genome, occupying about 11% of genomic space
[20]. Because gene conversion also acts on relatively short sequences (beginning with 10 nt
[17]), not only are entire
Alu sequences substituted, but also partial
Alu–
Alu gene conversion occurs, resulting in hybrid elements (e.g., hybrids with 5′-
AluS and 3′-
AluY
[21][22], ).
Changing the TE type via gene conversion might impact the global genome architecture and, for genome scientists, may also lead to faulty genome annotations and obstruct TE-based phylogenetic reconstructions. The phylogenetic presence of identical orthologous TE elements in several species indicates their close relationships, the identification of which can be compromised if gene conversion results in altered element types. We previously showed that parallel insertions and precise deletions of
Alus are rare in primates, confirming their usefulness as virtually homoplasy-free markers in phylogenetic studies
[23]. However, no study has yet evaluated gene conversion as an additional possible source of confounding TE presence/absence patterns. Replacing one
Alu type with another in a monophyletic species group can lead to an incorrect conclusion about their phylogenomic relationship. Therefore, to determine the extent of possible homoplasy caused by gene conversion and the frequency of gene conversion in TEs of different ages, we performed a systematic screening for gene conversion among
Alu elements belonging to clearly different primate
Alu subfamilies and types (
AluY/
AluS and
AluY/
AluYc).
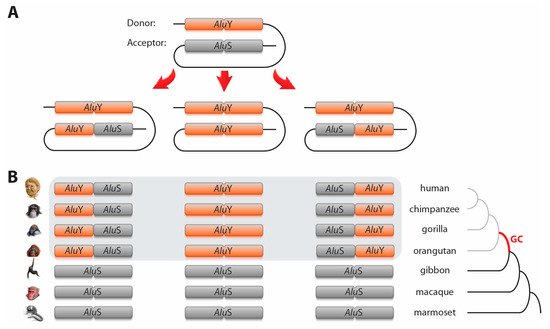
2. Alu–Alu element type change via gene conversions
Here we present for the first time a systematic, genome-wide screening of primate genomes for clear
Alu–
Alu element type change via gene conversions. Two recently developed tools were combined to find 98 specific cases of gene conversion. fastCOEX derived
Alu loci with almost TE-free flanks, and 2-n-way extracted their orthologous sequences in various primate species. Gene conversion is identifiable when different
Alu subfamilies or types recombine (e.g.,
AluS change to
AluY or vice versa). From a RepeatMasker report of the human genome using fastCOEX
[24], we extracted human coordinates of 55,408
AluS and 12,689
AluY/Yc full-length elements with flanking regions largely free of other repetitive sequences. However, restricting our screening to these most reliable cases of
Alu TEs reduces the total dataset of human
Alus (~800.000 for
AluS plus
AluY) by about a tenth. We used the 2-n-way computer suite to retrieve 46,285 targeted
AluS and 8099
AluY/Yc elements orthologous loci for a set of hominoid species (see Section 2.2 under Methods). We then applied a local RepeatMasker analysis to both annotate each hominoid insertion at orthologous positions, and, in a search for human hybrid elements, to compare the element subtypes of 5′- and 3′-
Alu monomers for each human
Alu. After manual inspection to verify orthology, we identified 98 cases in which some primate species or species groups contained different
Alu elements or hybrid
Alus compared to the others in the group (Table 1, Supplementary Table S1, and Supplementary File S2). It has to be mentioned that this number underestimates the actual extent of
Alu gene converted loci. The more similar elements are, the more probable they involve in gene conversion. However, gene conversion of identical elements is difficult to trace. About half of the identified gene converted
Alu elements were located in gene regions (introns or UTRs). The other half was found in the intergenic areas of the genome (Supplementary Table S1). However, because of restricting our survey to
Alus free from flanking TEs, we underestimate the portion of
Alu–
Alu gene conversion in intergenic regions.
Table 1. Gene conversion cases among Alu elements.
| Donor |
Acceptor |
Replaced Part of Alu |
Number of Loci |
| Gene conversion with identified direction |
| AluY |
AluS |
Complete Alu |
11 |
| AluY |
AluS |
3′-Alu unit (S-Y hybrid) |
23 |
| AluY |
AluS |
5′-Alu unit (Y-S hybrid) |
6 |
| AluS |
AluY |
Complete Alu |
3 |
| AluS |
AluY |
3′-Alu unit (Y-S hybrid) |
6 |
| AluS |
AluY |
5′-Alu unit (S-Y hybrid) |
3 |
| AluYc |
AluY |
Diagnostic indel |
9 |
| AluY |
AluYc |
Diagnostic indel |
3 |
| Gene conversion with unidentified direction |
| Unidentified |
Unidentified |
AluS–AluY hybrid |
31 |
| Complex scenario |
Complex scenario |
Multiple gene conversion |
3 |
For 64 of the 98 gene conversion loci, we were able to reconstruct the original ancestral
Alu element type and to determine the direction of gene conversion. For the
AluS to
AluY conversions (40 loci; Table 1, first three lines), the older
AluS elements (
AluSs ceased their main activity before the diversification of Catarrhini) were replaced by younger, potentially active
AluY elements (
AluYs exhibited their main activity starting with the divergence of Catarrhini)
[25]. This suggested that young, actively transcribed DNA regions were the preferred donors for gene conversion
[26]. However, we also observed incidences in which the reverse process occurred (12 cases; Table 1, line 4–6), providing evidence that old inactive elements might replace young active elements via gene conversion resulting in a sort of “life after death” spreading throughout the genome after silencing. For 14 of the reconstructed 52 loci involving both
AluS and
AluY elements, we detected gene conversion of the complete acceptor element, whereas in the remaining 38 loci only partial gene conversion occurred, leading to “mosaic” or hybrid elements (e.g., a hybrid of
AluS 5′-monomer and
AluY 3′-monomer). It should be mentioned that
AluY/
AluS gene conversion events resulted in hybrids of
AluY 5′-monomer and
AluS 3′-monomer (12 cases) can also be potentially
AluSc8/
AluS gene conversion because the 5′-monomer of
AluSc8 shares the diagnostic mutations of
AluY and the 3′-monomer of
AluS. Furthermore, we detected an additional 31 cases of hybrid elements, in which we were unable to assign the pre-conversional state of the
Alu elements (Table 1, line 9). We were unable to categorize
AluY/
AluS hybrids for cases of unidentified ancestral origins because they were indistinguishable from
AluSc8 elements. We also observed 12 incidences of gene conversion among
AluY and
AluYc elements and 3 cases, in which the
Alu loci underwent more than one gene conversion event during primate evolution (Table 1).
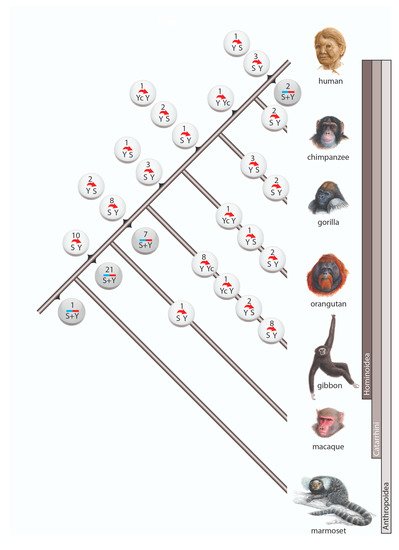
Among the 98 cases of gene conversion, 64 occurred on the lineage leading to humans (including 6 instances after human split from chimpanzee), whereas 31 gene conversions occurred on the terminal branches of other investigated primates (Figure 2). Within Anthropoidea we distinguished three waves of high gene conversion events: (1) on the ancestral branch of Catarrhini (31 conversions), (2) on the ancestral branch of hominoids (17 conversions), and (3) in the gibbon lineage (19 conversions). The first two of these higher incidences might be explained by the longer lengths of the ancestral internodes leading to Catarrhini and hominoids, both leaving substantial times for the occurrence and fixation of gene conversion events. The increased gene conversion events in gibbons might be partially explained by the more highly active gibbon-specific
AluY elements (
AluYd3a1_gib
[27]), which contain the same diagnostic deletion as the
AluYc element.
Figure 2. Gene conversion in primates. Circles represent incidences of gene conversion including the number of such occurrences and their direction. White circles are incidences in which the ancestral Alu element and the direction of conversion were reconstructed. Gray circles represent incidences with unidentifiable conversion direction. The 3 cases with complex scenarios are not shown.
Another gene conversion-rich branch was that leading to gorilla. In our initial analysis, we screened the gorGor4 genome assembly (gorilla Kamilah, UCSC https://genome-euro.ucsc.edu/cgi-bin/hgGateway, accessed on 9 June 2021) and found 12 gene conversions (Supplementary File S3). A previous examination of interlocus gene conversion in gorGor4
[28], also observed a more frequent occurrence of gene conversion in gorilla than in other great apes. However, our expanded analysis of another gorilla genome (gorilla Susie, gorGor5) revealed only 5 gene conversion events (Figure 2, Supplementary Table S1), suggesting that the difference between gorGor4 and gorGor5 is an individual variation or a genomic artifact of the gorGor4 assembly. The gorGor6 assembly (August 2019, assembly Kamilah_GGO_v0/gorGor6) that recently became available carries none of the previously detected cases of gene conversions found solely in gorGor4, suggesting there might be assembly errors in gorGor4. Learning from gorilla, we compared gene conversion patterns for at least two related species or independent assemblies in cases when gene conversion occurred on a terminal branch to avoid such assembly issues.
We conducted a population analysis of human-specific gene conversions (6 cases), including 35 human individual genomes from Africa, Asia, America, and Europe (Supplementary Table S2). We found a consistent gene conversion pattern in all investigated genomes for the 5 loci containing AluS to AluY conversions. For the remaining one locus (AluY converted to AluS), gene conversion was only detected in some human individuals. Contrary to our expectations, we could not find a phylogenetic pattern of the gene conversion distribution among 35 individuals. Orthologous Alu gene conversion was found in 2 of 9 African individuals, 5 of 13 Asian, 1 of 4 American, 1 of 1 Puerto Rican, and 5 of 8 European individuals. We suggest that such a mosaic of gene conversion events might result from duplication of Alu loci in the human genome with the subsequent gene conversion in one of the copies. Alternatively, multiple independent conversions could have occurred.
In the present study, we examined gene conversion events leading to changes in the Alu subfamily or type affiliations in selected hominoids. It should be noted that because of sequence similarity, gene conversion occurs most frequently among identical or closely related elements, and is then unrecognizable. Here we showed that AluS/Y/Yc gene conversion occurred in all hominoid lineages. We suggest that the observed patterns of Alu–Alu gene conversion in hominoids are also representative of other primate species and TE types.
Parallel insertion, exact deletion, or gene conversion might lead to apparently conflicting presence/absence patterns at orthologous loci. Doronina et al.
[23] showed there to be a negligibly low frequency of conflicting phylogenetic signals amongst
Alu elements in primates. However, they did not examine gene conversion. Although Aleshin et al.
[8] found a notable quantity of potential
Alu–
Alu gene conversions, their screening method (ignoring diagnostic
Alu positions) does not evaluate the contribution of gene conversion to homoplasy. Similar to the data in Doronina et al.
[23], we estimate the frequency of gene conversion-related homoplasy in the human-chimpanzee-rhesus macaque model group to be 0.0006% in human (3/544,034 × 100%) and 0.0004% in chimpanzee (2/544,034 × 100%), where 544,034 is the number of
Alu insertions present in the Catarrhini ancestral lineage. Thus, we provide evidence for the existence of homoplasy caused by gene conversion, but show that the frequency is even lower than parallel insertions or precise deletions.
It should be mentioned that the classical, distance-based (the divergence of a TE sequence from a consensus sequence) calculations of the ages of TEs used in evolutionary studies might be distorted by gene conversion
[9][11][29]. Our results suggest that transposition-in-transposition-based analyses
[25] that take into account element types rather than accumulated mutations in TE sequences may provide a more reliable alternative. Indeed, we detected relatively few gene conversion events per lineage affecting diagnostic positions that resulted in TE subfamily or type changes, whereas the sharing of non-diagnostic mutations among
Alus via gene conversion was shown to be a frequent phenomenon
[8].
In summary, the footprints of gene conversion are directly detectable by genome-wide comparisons of deviating annotations of orthologous TEs in different species (e.g., orthologous Alu SINEs with different subfamily or type affiliations in primates). Many potential incidences of partial gene conversion were detected that resulted in hybrid elements. Incidences of gene conversion in TEs are frequent enough to visualize by genome-level screenings but rare enough that they do not challenge large-scale phylogenetic TE presence/absence studies.