| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Byung-Hoon Jeong | + 1878 word(s) | 1878 | 2020-06-11 08:37:13 |
Video Upload Options
Transmissible spongiform encephalopathies (TSEs) have been reported in a wide range of species. However, TSE infection in natural cases has never been reported in dogs. Previous studies have reported that polymorphisms of the prion protein gene (PRNP) have a direct impact on the susceptibility of TSE. However, studies on polymorphisms of the canine PRNP gene are very rare in dogs. We examined the genotype, allele, and haplotype frequencies of canine PRNP in 204 dogs and analyzed linkage disequilibrium (LD). In addition, to evaluate the impact of nonsynonymous polymorphisms on prion protein (PrP), we carried out in silico analysis.
Abstract: Transmissible spongiform encephalopathies (TSEs) have been reported in a wide range of species. However, TSE infection in natural cases has never been reported in dogs. Previous studies have reported that polymorphisms of the prion protein gene (PRNP) have a direct impact on the susceptibility of TSE. However, studies on polymorphisms of the canine PRNP gene are very rare in dogs. We examined the genotype, allele, and haplotype frequencies of canine PRNP in 204 dogs and analyzed linkage disequilibrium (LD). In addition, to evaluate the impact of nonsynonymous polymorphisms on prion protein (PrP), we carried out in silico analysis.
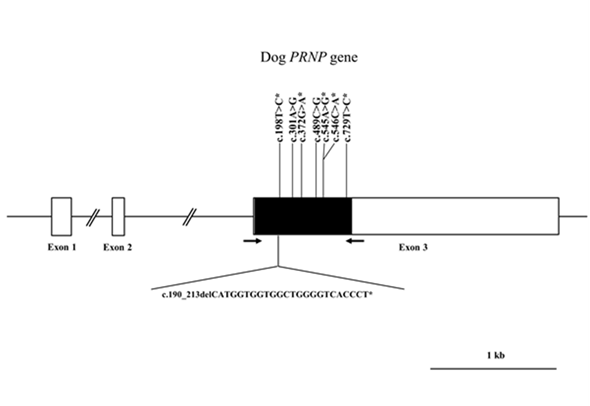
Figure 1. Gene map of polymorphisms identified in the canine prion protein (PRNP) gene on chromosome 24. The open reading frame (ORF) is indicated by a shaded block, and the 5ʹ and 3ʹ untranslated regions (UTRs) are indicated by white blocks. Arrows indicate the regions sequenced. The Y-shaped bar indicates the octapeptide deletion polymorphisms identified in the canine PRNP gene. Asterisks indicate the novel polymorphisms found in this study.
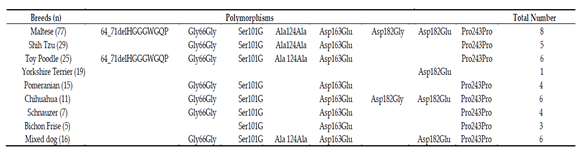
Table 1. Different distributions of PRNP polymorphisms in 8 dog breeds.
Transmissible spongiform encephalopathies (TSEs), also known as prion diseases, are neurodegenerative diseases caused by conversion of the normal prion protein (PrPC) into aggregated, self-propagating and disease-associated isoforms (PrPSc). TSE has been reported in a wide range of species, such as goats, sheep, cattle, mink, cats and humans [1-11]. However, during the outbreak of bovine spongiform encephalopathy (BSE) in the UK, BSE transmitted to cats through contaminated food. Although dogs were equally likely to have been exposed to BSE contaminated food, TSE infection was never reported in dogs [12, 13]. Since polymorphism of the PRNP gene has been associated with the susceptibility to prion diseases [5, 11, 14, 15], we amplified the ORF region of the canine PRNP gene to identify the genetic polymorphism of this gene. We identified a total of eight polymorphisms, including two novel nonsynonymous SNPs and one insertion/deletion (Figure 1). We identified strong LDs and six major haplotypes among eight polymorphisms. The distribution of haplotypes was significantly different among the eight dog breeds. In addition, the number of identified polymorphisms was different from each dog breed (Table 1). Notably, Yorkshire Terrier showed the lowest number of polymorphisms in dog breeds with more than 12 samples capable of excavating 1% frequencies of SNPs with 96% probability (Table 1). Since the wolf and dog PrPs have the same amino acid sequence, the evolutionary distance of the PRNP gene between dog and wolf can be estimated according to the number of polymorphisms. In comparison with Maltese, Shih Tzu, Toy Poodle, and Pomeranian, which showed highly polymorphic PRNP gene, Yorkshire Terrier is presumed to be a close evolutionary distance of the PRNP gene with wolf.
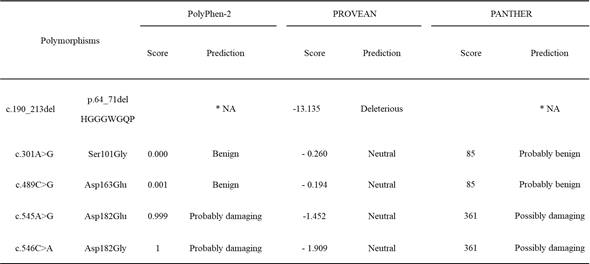
Table 2. In silico analysis of nonsynonymous SNPs of the PRNP gene in dogs.
We also estimated the impact of polymorphisms on dog PrP using PolyPhen-2, PROVEAN, and PANTHER. All three in silico programs predicted that Asp163Glu was benign. A previous study reported that Asp163Glu did not influence the susceptibility to TSE of transgenic mice expressing dog-specific amino acids 158Asp and 158Glu [16]. Codon 158 in mouse PrP is equivalent to codon 163 in dog PrP. In the present study, we observed similar results using in silico programs in which Asp163Glu does not impact the structure and/or function of dog PrP. Notably, PROVEAN and PANTHER predicted that the p.64_71del HGGGWGQP, Asp182Gly, and Asp182Glu polymorphisms can impact the function and/or structure of dog PrP (Table 2). These estimations suggested the possibility that p.64_71del HGGGWGQP, Asp182Gly, and Asp182Glu can impact the susceptibility of dogs to TSE (Table 2). However, Asp182Gly and Asp182Glu were predicted as neutral using PROVEAN. Because PROVEAN was estimated using clustering of basic local alignment search tool (BLAST) and comparing homologs collected from a database, PROVEAN predicted that Asp182Gly and Asp182Glu did not impact the function of PrP.
We also estimated the impact of polymorphisms on dog PrP using PolyPhen-2, PROVEAN, and PANTHER. All three in silico programs predicted that Asp163Glu was benign. A previous study reported that Asp163Glu did not influence the susceptibility to TSE of transgenic mice expressing dog-specific amino acids 158Asp and 158Glu [16]. Codon 158 in mouse PrP is equivalent to codon 163 in dog PrP. In the present study, we observed similar results using in silico programs in which Asp163Glu does not impact the structure and/or function of dog PrP. Notably, PROVEAN and PANTHER predicted that the p.64_71del HGGGWGQP, Asp182Gly, and Asp182Glu polymorphisms can impact the function and/or structure of dog PrP (Table 2). These estimations suggested the possibility that p.64_71del HGGGWGQP, Asp182Gly, and Asp182Glu can impact the susceptibility of dogs to TSE (Table 2). However, Asp182Gly and Asp182Glu were predicted as neutral using PROVEAN. Because PROVEAN was estimated using clustering of basic local alignment search tool (BLAST) and comparing homologs collected from a database, PROVEAN predicted that Asp182Gly and Asp182Glu did not impact the function of PrP.
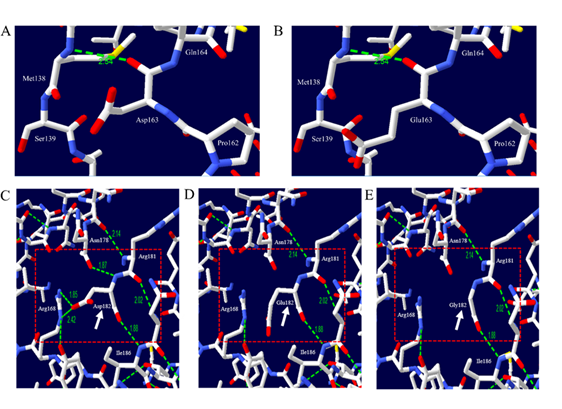
Figure 2. Prediction of 3D structure and hydrogen bonds of dog prion protein (PrP). The white arrow indicates target amino acid residues. The red box indicates adjacent amino acid residues. The green dotted line indicates hydrogen bonds. The green numbers indicate the distance of the hydrogen bonds. (A) 3D structure of dog PrP with allele Asp163, (B) 3D structure of dog PrP with allele Glu163, (C) 3D structure of dog PrP with allele Asp182, (D) 3D structure of dog PrP with allele Glu182, and (E) 3D structure of dog PrP with allele Gly182.
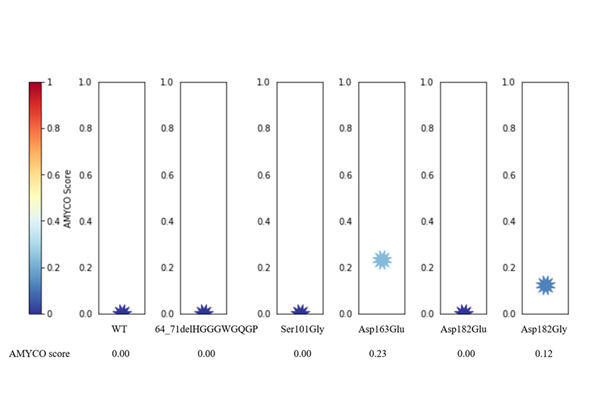
Figure 3. Prediction of aggregation propensity according to alleles of dog prion protein (PrP) polymorphisms. The impact of polymorphisms on the aggregation propensity of dog PrP was evaluated as values from 0.0 to 1.0 by AMYCO analysis. AMYCO scores <0.45 and >0.78 indicate low and high aggregation propensities of the protein, respectively.
Next, we predicted the 3D structure of dog PrP to evaluate the impact of three nonsynonymous SNPs, including Asp163Glu, Asp182Glu, and Asp182Gly. We compared the distribution of hydrogen bonds between alleles Asp163 and Glu163 of dog PrP. The distribution of hydrogen bonds in dog PrP is identical between the Asp163 and Glu163 alleles (Figure 2 A,B). Dog PrP with Asp182 was predicted to have four hydrogen bonds. However, dog PrP with Glu182 and Gly182 was predicted to have only one hydrogen bond (Figure 2 C,D). The number of hydrogen bonds can affect the stability and structure of proteins [17-19]. Because the stability of PrP is related to the susceptibility of prion disease, Asp182Glu and Asp182Gly SNPs of the canine PRNP gene can influence the susceptibility to TSE of dogs. We estimated the impact of the polymorphism of the canine PRNP gene on the aggregation propensity of dog PrP and found that dog PrP with Asp163Gly and Asp182Gly (score 0.12) had a higher aggregation propensity than that of wild-type dog PrP (Figure 3). Collectively, Asp182Glu, and Asp182Gly are presumed to be deleterious. Based on our analysis, Shih Tzu, Toy Poodle, and Pomeranian, which do not carry Asp182Glu and Asp182Gly, are presumed to be resistant to prion disease compared to Maltese and Yorkshire Terrier in dog breeds with more than 12 samples. It indicates that evolutionary sensitization to prion infection can be occurred in Maltese and Yorkshire Terrier. To confirm the impact of Asp182Glu and Asp182Gly SNPs on the susceptibility to prion disease of dogs, infection experiments with prion agents will be necessary in MDCK cells and transgenic mice expressing dog PrP with two amino acid substitutions, Asp182Glu and Asp182Gly, in the future.
Although most of our analysis has been focused on nonsynonymous SNPs, there are recent evidences that synonymous SNPs introduce less commonly used codons, which may alter the speed of translation and ultimately folding, function, and stability of the mature protein [20, 21]. Since prion diseases are induced by misfolded prion protein, these are important considerations of synonymous SNPs. Further study of synonymous SNP is highly desirable in the future.
The article has been published on https://doi.org/10.3390/ijms21114160
- McIntyre, K.M., et al., Epidemiological characteristics of classical scrapie outbreaks in 30 sheep flocks in the United Kingdom. PLoS One, 2008. 3(12): p. e3994.
- Curcio, L., et al., A review on classical and atypical scrapie in caprine: Prion protein gene polymorphisms and their role in the disease. animal, 2016. 10(10): p. 1585-1593.
- Sheridan, H.A., et al., A temporal-spatial analysis of bovine spongiform encephalopathy in Irish cattle herds, from 1996 to 2000. Canadian Journal of Veterinary Research, 2005. 69(1): p. 19.
- Imran, M. and S. Mahmood, An overview of animal prion diseases. Virology journal, 2011. 8(1): p. 493.
- Jeong, B.H. and Y.S. Kim, Genetic studies in human prion diseases. J Korean Med Sci, 2014. 29(5): p. 623-32.
- Mathiason, C.K., et al., Susceptibility of domestic cats to chronic wasting disease. J Virol, 2013. 87(4): p. 1947-56.
- Kim, S.-K., et al., Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Scientific Reports, 2019. 9(1): p. 1-10.
- Jeong, B.H., et al., Bovine spongiform encephalopathy (BSE)‐associated polymorphisms of the prion protein (PRNP) gene in K orean native cattle. Animal genetics, 2013. 44(3): p. 356-357.
- Kim, Y.-C. and B.-H. Jeong, Bovine spongiform encephalopathy (BSE) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. Journal of Dairy Research, 2018. 85(1): p. 7-11.
- Kim, Y.-C. and B.-H. Jeong, The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta Veterinaria Hungarica, 2017. 65(2): p. 291-300.
- Baylis, M. and W. Goldmann, The genetics of scrapie in sheep and goats. Current molecular medicine, 2004. 4(4): p. 385-396.
- Collee, J.G. and R. Bradley, BSE: a decade on-part 2. The Lancet, 1997. 349(9053): p. 715-721.
- Collee, J.G. and R. Bradley, BSE: a decade on—Part I. The Lancet, 1997. 349(9052): p. 636-641.
- Murdoch, B.M. and G.K. Murdoch, Genetics of prion disease in cattle. Bioinformatics and biology insights, 2015. 9: p. BBI. S29678.
- Lloyd, S., S. Mead, and J. Collinge, Genetics of prion disease, in Prion Proteins. 2011, Springer. p. 1-22.
- Vidal, E., et al., Dogs are resistant to prion infection, due to the presence of aspartic or glutamic acid at position 163 of their prion protein. The FASEB Journal, 2020. 34(3): p. 3969-3982.
- Myers, J.K. and C.N. Pace, Hydrogen bonding stabilizes globular proteins. Biophys J, 1996. 71(4): p. 2033-9.
- Marqusee, S. and R.T. Sauer, Contributions of a hydrogen bond/salt bridge network to the stability of secondary and tertiary structure in λ repressor. Protein Science, 1994. 3(12): p. 2217-2225.
- Alber, T., et al., Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature, 1987. 330(6143): p. 41.
- Im, E.-H. and S.S. Choi, Synonymous codon usage controls various molecular aspects. Genomics & informatics, 2017. 15(4): p. 123.
- Fung, K.L., et al., MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer research, 2014. 74(2): p. 598-608.