Here, we report the functional characterization of the MLO4 protein in Arabidopsis roots. The MLO4 was identified as interacting with CML12 in a screening for the interaction between the proteins from Arabidopsis MLO and calmodulin/calmodulin-like (CaM/CML) families using yeast two hybrid (Y2H) assays. Then, the interaction between MLO4 and CML12 was further verified by Luciferase Complementation Imaging (LCI) and Bimolecular Fluorescence Complementation (BiFC) assays. Genetic analysis showed that the mlo4, cml12, and mlo4 cml12 mutants displayed similar defects in root gravity response. These results imply that the MLO4 might play an important role in root gravity response through interaction with CML12. Moreover, our results also demonstrated that the interaction between the MLO and CaM/CML families might be conservative.
1. Introduction
Mildew resistance locus O (MLO) was first identified as being resistant against powdery mildew (PM) infection in barley
[1]. Then, the MLO proteins were found in many plant species.
Arabidopsis has at least 15 MLO proteins that diverged into several clades and were involved in different physiological processes, such as sexual reproduction, PM infection, and root thigmomorphogenesis
[2][3][4][5]. MLO7/NORTIA (NTA), a downstream component of the receptor-like kinase FERONIA (FER), is involved in pollen tube reception and PM infection
[6]. Study showed that its function is dependent on NTA homooligomerization and its carboxyl-terminal tail identity
[7]. MLO5/9/15 played roles in pollen tube responses to ovular signals, and MLO5/9 selectively recruit Ca
2+ channel CNGC18-containing vesicles to the plasma membrane
[8]. The MLO1, MLO2, MLO3, MLO6, MLO8, MLO9, MLO10, and MLO13 are expressed in discrete domains during reproductive development
[4]. The MLO2, MLO4, and MLO6 modulate defense responses against PM fungi and a number of other phytopathogens
[9]. Furthermore, the MLO2 also function as a negative regulator in plant ROS responses involving biotic and abiotic stress
[10]. Mutations in
MLO4 and
MLO11 exhibit abnormal root thigmomorphogenesis and gravity sensitivity
[3][11]. Genetical complementation with MLO4 domains indicate that the C-terminal cytoplasmic domain of MLO4 is necessary for regulation of asymmetrical root growth
[11].
All MLO proteins are predicted to share a conserved calmodulin-binding domain (CaMBD) in C-terminal cytoplasmic tail
[12][13]. The gel overlay assays demonstrated in vitro Ca
2+-dependent binding of CaM to the CaMBD of the MLOs from barley
[14]. However, little is known about the interaction of individual MLO with the specific calmodulins (CaMs) or calmodulin-like proteins (CML) in
Arabidopsis or other plants.
The
Arabidopsis genome encodes 7 CaMs and 50 CMLs, which are presumed to sense and transduce Ca
2+ signals
[15]. Functional studies of CaM/CML in plants revealed that this protein family converts calcium signals into transcriptional responses, protein phosphorylation, or metabolic changes to regulate plant development responding to the ever-changing environment
[16]. Some CaM/CML proteins are involved in plant growth and development, such as AtCaM7 regulating light-induced seedling development
[17][18], AtCaM2 functioning in pollen germination
[19], AtCML42 for trichome morphogenesis
[20], and AtCML23 and AtCML24 in flowering
[21][22]. Some CaM/CML proteins are indispensable for plants to respond to abiotic and biotic stress. The mutations in
AtCaM3 lead to reduction in thermotolerance of the mutant plants
[23]. AtCaM4 negatively regulates freezing tolerance by interacting with CaM-binding protein PATL1 in a CBF-independent manner
[24]. A knockout mutant of
AtCML9 enhances its tolerance to drought and salinity stress
[25]. AtCML20 is a negative regulator in guard cell ABA signaling during drought tolerance
[26]. AtCML8 and AtCML9 positively regulate plant immunity in response to
Pst inoculation
[27]. Less NtCaM13 expression causes more susceptibility to virulent bacteria and fungi in tobacco
[28]. Overexpression of pepper CaM1 and AtCML43 confers enhanced resistance to pathogens
[29]. All these data suggest that the CaM/CML-mediated defense signaling pathways involve a complex regulatory network.
Both MLO and CaM/CML proteins are important for plants. They have potential to interact with each other in between the two families through CaMBD
[12]. However, for now, it is just predicted by physical structure of the proteins, and not solid evidence, to prove their interaction or even their related biological function.
2. The Expression Patterns of MLO and CaM/CML Genes
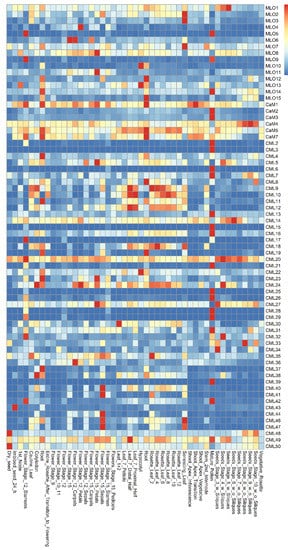
The co-expression patterns of MLO and CaM/CML genes were analyzed using the e-FP data from TAIR website (www.arabidopsis.org, accessed on 19 May 2021) (Figure 1). The results showed that the MLO and CaM/CML genes are expressed in various Arabidopsis tissues. The MLO1 was expressed more highly in seeds than in other tissues. The CaM/CML genes, which exhibit similar expression in seeds, included 12 members, namely, CaM4, CML14, CML20, CML24, CML27, CML32, CML33, CML34, CML43, CML48, CML49, and CML50. The MLO5, MLO9, and MLO14 genes were highly expressed in pollen. In the pollen, 17 CaM/CML genes, including CaM2, CaM3, CML2, CML3, CML6, CML7, CML13, CML15, CML16, CML21, CML26, CML28, CML29, CML31, CML39, CML42, and CML49, were found to be highly co-expressed. In flowers, four MLO genes (MLO4, MLO6, MLO8, and MLO11) and nine CML genes (CML5, CML24, CML27, CML36, CML40, CML41, CML44, CML46, and CML47) were highly co-expressed. The four MLO genes, MLO2, MLO3, and MLO8, were found to be highly expressed in leaves; similarly, five CML genes were highly expressed in leaves, namely, CML9, CML10, CML18, CML35, and CML46. In roots, MLO12 gene was highly co-expressed, together with five CaM/CML genes (CaM1, CaM5, CaM7, CML8, and CML19). MLO4 and CML12 were expressed in many tissues, including roots. Although the highest expression levels of both MLO4 and CML12 did not present in roots, they were highly co-expressed in roots, implying both of them may function in roots.
Figure 1. Expression pattern of Arabidopsis MLO and CaM/CML family genes.
3. Screening for Interactive Pairs of MLO and CaM/CML Proteins
The MLO proteins contain a CaMBD in the C-terminus region
[12]. Therefore, the C-terminus regions of the 15 MLO proteins were used to characterize the interaction of MLO and CaM/CML proteins by Y2H. The results showed that each MLO protein may interact with at least one CML protein in yeast cells (Table 1). In particular, the NTA/MLO7, which was involved in pollen tube reception, could interact with up to 11 candidate CML proteins, including CML8-12, CML26, CML30, CML37, CML38, CML40, and CML40. Similar results were also obtained from the assays for the three closely-related proteins MLO4, MLO11, and MLO14, which function in root thigmomorphogenesis
[11]. MLO4 interacted with CML12, CML40, and CML44. MLO11 interacted with CML47. MLO14 interacted with CML17, CML18, and CML49.
Table 1. Interactive protein pairs between MLO and CaM/CML family.
| MLOs |
CaM/CML Proteins that Interacted with Corresponding MLO Proteins |
| MLO1C |
CaM2, CML29 |
| MLO2C |
CML9, CML18 |
| MLO3C |
CML18, CML20, CML23, CML26, CML32, CML40, CML44 |
| MLO4C |
CML12, CML40, CML44 |
| MLO5C |
CML18, CML26, CML40, CML44 |
| MLO6C |
CML8, CML10, CML20, CML26, CML73, CML40, CML44, CML49 |
| MLO7C |
CML8, CML9, CML10, CML11, CML12, CML26, CML30, CML37, CML38, CML40, CML44 |
| MLO8C |
CML8, CML9, CML23, CML26, CML37, CML40, CML44 |
| MLO9C |
CML10, CML42, CML44, CML49 |
| MLO10C |
CML8, CML9, CML10, CML26, CML37, CML44 |
| MLO11C |
CML47 |
| MLO12C |
CML10, CML12, CML30, CML35, CML36, CML37, CML44 |
| MLO13C |
CML32, CML40, CML41 |
| MLO14C |
CML17, CML18, CML49 |
| MLO15C |
CML10, CML44 |
4. LCI and BiFC Assays Further Demonstrated That MLO4 Interacted with CML12
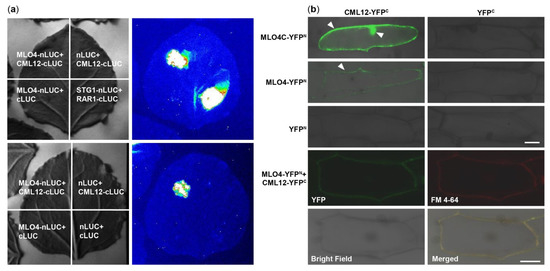
At first, MLO4 and CML12 were chosen to verify the results of Y2H screening. Then, the LCI results showed that CML12 interacted with the full length of MLO4 in tobacco cells (Figure 2a). BiFC results demonstrated that the CML12 interacted with the cytoplasmic domain of MLO4, in concert to Y2H result. Furthermore, when the full length of MLO4 was used for the assays, the interactive signal was detected only on the plasma membrane, which was co-localized with the staining signals of the membrane dye FM 4-64 (Figure 2b). This result was consistent to the ones obtained from the membrane localization of MLO4 with a signal peptide and seven transmembrane domains
[11].
Figure 2. CML12 interacted with MLO4. (a) LCI results showed CML12 interacted with full length of MLO4 in tobacco cells. (b) BiFC results showed that CML12 interacted with cytoplasmic domain of MLO4 in onion cells, and the signal was detected in nucleus and periphery. When CML12 interacted with full length of MLO4, the signal was only detected on the plasma membrane, colocalized with FM 4-64. The YFP signals are indicated by pointed triangles in each figure. Bars = 50 μm.
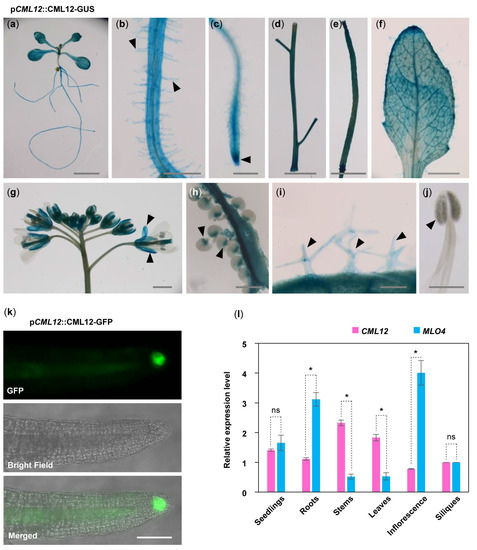
5. MLO4 and CML12 Displayed a Similar Expression Pattern
To understand the relationship between
MLO4 and
CML12, we further investigated whether
MLO4 and
CML12 have a co-expression pattern in vivo. First, GUS staining of p
CML12::CML12-GUS transgenic lines showed that
CML12 was expressed in most of the vegetable tissues, such as seedlings, roots, root hairs, leaves, trichomes, stems, siliques, calyxes, and chalazal end of ovules but not in pollen (Figure 3a–j). In addition, p
CML12::CML12-GFP transgenic lines showed that the CML12-GFP fusion protein was detected in the root cap (Figure 3k). Real-time PCR results showed that
MLO4 and
CML12 shared a similar expression pattern. Both genes were expressed constitutively in many tissues, including seedlings, roots, stems, leaves, inflorescence, and siliques. Expression levels of both genes were closely similar in seedlings.
MLO4 was expressed with twice the
CML12 expression in roots (Figure 3l). A previous study revealed that the p
MLO4::MLO4-GFP was also expressed in the roots, and MLO4-GFP fusion protein was detected in the root epidermal cell
[11]. Therefore, both CML12 and MLO4 proteins were expressed in roots, suggesting that they may function coordinately.
Figure 3. Expression pattern of CML12 Gene. GUS staining of pCML12::CML12-GUS transgenetic lines showed CML12 was expressed in most of the vegetable tissues: Seedling (a), root hairs (b), root tip (c), stems (d), silique (e), leaf (f), sepals (g), chalazal end of ovules (h), and trichomes (i), not in pollen (j), as indicated by pointed triangles in each figure. Bars = 100 μm. (k) pCML12::CML12-GFP transgenetic lines showed CML12-GFP was detected in root cap. Bar = 20 μm. (l) Real-time PCR showed that CML12 and MLO4 genes both were expressed in various tissues of Arabidopsis. ns: not significant; the * above bars indicate the t-test results at significant level of 0.05.
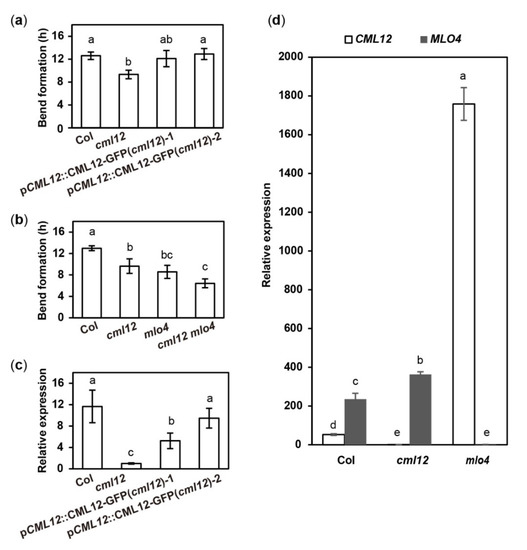
6. The mlo4 and cml12 Mutants Shared Similar Phenotypes
The MLO4 has been characterized as regulating root thigmomorphogenesis, including gravitropism in roots
[11]. Previous characterization of
cml12 revealed that the mutations in
CML12 exhibited a similar phenotype
[30]. To study the genetic relationship between
MLO4 and
CML12, we further characterized their roles in root growth.
The
cml12-3 mutant (SALK_122731) was obtained from Dr. Sherryl R. Bisgrove (Department of Biological Sciences, Simon Fraser University, Burnaby, BC, Canada). Real-time PCR showed that
cml12-3 was a knock-out mutant, as reported by Gleeson et al.
[31]. To test its gravity response,
cml12-3 seedlings were vertically grown on the agar minimal medium (0.25% sucrose) for 7 d (16 h light/8 h dark cycle); then, the plates were rotated by 90° in the clockwise direction. The roots bent down in response to gravity change until root tips became parallel with new gravity vector (Figure 4a). Compared to the time (12.60 ± 0.66 h) for the wild-type roots to form a bend, the
cml12-3 roots took less time (9.33 ± 0.75 h) to form a bend (Figure 5a), suggesting that
cml12-3 roots were more sensitive to gravity response.
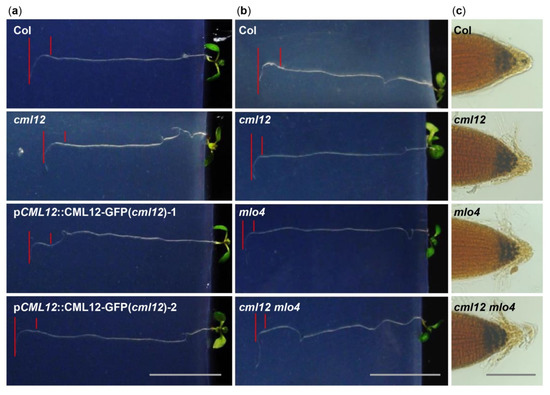
Figure 4. Phenotype analysis of cml12-3 and mlo4-5 mutants. cml12-3 and mlo4-5 mutants were indicated, respectively, as cml12 and mlo4, for short. (a) 7-day-old seedlings growing through the agar medium were rotated by 90° in the clockwise direction. Roots bent down to respond to gravity change. Red lines point to tip position at the time of rotation and the location where root growth became parallel with new gravity vector. Bar = 1 cm. (b) Roots bent down to respond to gravity change. Red lines point to tip position at the time of rotation and the location where root growth became parallel with new gravity vector. Bar = 1 cm. (c) Starch was detected using KI-I2 stain Starch in single mutants cml12-3 and mlo4-5 and double mutant cml12-3 mlo4-5 was obviously more than that in wild-type root tips. Bar = 100 μm.
Figure 5. Quantitative analysis of bend formation and gene expression in cml12-3 and mlo4-5 mutants. cml12-3 and mlo4-5 mutants were indicated, respectively, as cml12 and mlo4 for short. (a)The time taken to form a bend in Col, cml12-3, pCML12::CML12-GFP (cml12-3)-1, and pCML12::CML12-GFP (cml12-3)-2 were 12.60 ± 0.66, 9.33 ± 0.75, 12.10 ± 1.42, and 12.90 ± 0.96 hours, respectively, 80 < n < 150. (b) The time taken to form a bend in Col, cml12-3, mlo4-5, and cml12-3 mlo4-5 were 12.97 ± 0.45, 9.63 ± 1.36, 8.57 ± 1.22, and 6.43 ± 0.83 h, respectively, 80 < n < 150. (c) Real-time PCR showed that expression of CML12 gene was restored in pCML12::CML12-GFP (cml12-3/-) plants. (d) Real-time PCR showed that relative expression of CML12 increased significantly in mlo4-5 mutant. Different letters above the bars indicate significant differences (P < 0.05, Bonferroni correction) in a pairwise comparison using the least significant difference (LSD) method.
To verify the phenotype of cml12-3 in gravity response, a complementation assay was performed using the construct pCML12::CML12-GFP in PCAMBIA 1300 vector containing the native promoter (1500 bp upstream ATG) and coding sequence of CML12 (975 bp). Two pCML12::CML12-GFP transgenic cml12-3 mutant lines were chosen to further characterize their root phenotype. The average times for the roots from each pCML12::CML12-GFP (cml12-3/-) transgenic lines to form a bend were 12.10 ± 1.42 h and 12.90 ± 0.96 h, respectively, similar to that of wild-type roots (12.60 ± 0.66 h) (Figure 4a). The real-time PCR results showed that CML12 expression was higher in transgenic roots than that in cml12-3 mutant roots (Figure 5a). Therefore, CML12 was able to restore the defect of cml12-3 mutants in gravity response, further demonstrating that CML12 is involved in gravity response.
The previous study showed that
mlo4-1,
mlo4-3, and
mlo4-4 mutants exhibited different growth patterns on the reclined medium than wild-type
[11]. In this study, we identified another Salk allele
mlo4-5 in which the T-DNA was inserted in the last exon of
MLO4, different from the previously-reported alleles. Real-time PCR showed that
mlo4-5 was a knock-out mutant and expressed truncated protein. At first, an allelic analysis was performed. The F1 progeny of
mlo4-5 from the crosses with
mlo4-4 exhibited a resembled phenotype in growth pattern. The loops in each root of
mlo4-4/+; mlo4-5/+ plant were denser with shorter wavelengths, and its phenotype was similar to that of single mutant-
mlo4-4/- and
mlo4-5/-. The result demonstrated that
mlo4-5 also was an effective mutant of
MLO4 gene. Therefore, the
mlo4-5 was used for further characterization.
The phenotypic comparison in root gravity response was performed with the single mutant cml12-3 (cml12-3/-) and mlo4-5 (mlo4-5/-), as well as the double mutant cml12-3 mlo4-5 (cml12-3/-; mlo4-5/-). The time for bend formation was 12.97 ± 0.45 h in wild-type roots. In contrast, the roots of the single mutant cml12-3, mlo4-5 took 9.63 ± 0.45 h and 8.57 ± 1.22 h to form bends, respectively. The roots of the double mutant cml12-3 mlo4-5 took relatively shorter time (6.43 ± 0.83 h) to form a bend (Figure 4b, Figure 5b), indicating that the phenotype of double mutant was a little stronger than those of the single mutants.
Starch grains in root tips are sensors of gravity response
[30]. Therefore, the starch stain assays were further performed to compare the starch contents in the mutants and wild-type root tips using KI-I
2 staining. Contents of the starch grains in the single mutant
cml12-3 and
mlo4-5, as well as the double mutant
cml12-3 mlo4-5, were obviously higher than that in wild-type root tips (Figure 4c).
In addition, cml12-3, mlo4-5, and cml12-3 mlo4-5 mutant roots shared similar growth patterns with denser loops and shorter wavelengths on the reclined agar medium. The phenotype of the double mutant cml12-3 mlo4-5 was not obviously aggravated.
7. Mutation in MLO4 Affected the Expression of CML12
CaM/CMLs bound calcium and were supposed to regulate target proteins downstream
[16]. To further study the possible regulatory relationship between
CML12 and
MLO4, their relative expression levels in each other mutants were analyzed. The real-time PCR assay showed the expression of
CML12 was significantly elevated in
mlo4-5 mutant, far more than MLO4 expression changing in
cml12-3 mutant (Figure 5d). Thus,
MLO4 likely modulates the expression of
CML12.