
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pilar Domingo-Calap | + 2546 word(s) | 2546 | 2020-05-25 10:16:59 | | | |
| 2 | Nicole Yin | -2 word(s) | 2544 | 2020-06-16 03:50:40 | | | | |
| 3 | Nicole Yin | Meta information modification | 2544 | 2020-11-05 11:34:28 | | |
Video Upload Options
Biofilms are clusters of bacteria that live in association with surfaces, attached to other bacterial cells and to the surface by an extracellular polymeric matrix. Biofilms are capable of adhering to a wide variety of surfaces, both biotic and abiotic, including human tissues, medical devices, and other materials, representing a major threat causing infectious diseases and economic losses. Unfortunately, current antibiotics and common disinfectants have shown limited ability to remove biofilms adequately. Here, phage-based treatments are proposed as promising alternatives for biofilm eradication, including phage therapy, phage-derived enzymes, genetically modified phages, and phages in combination with antibiotics.
1. Introduction
Understanding the underlying mechanisms involved in phage resistance and the co-evolutionary interactions between phages and biofilms is very important to design phage-based treatments and to minimize the likelihood of resistance emergence[1]. Phage-based treatments include phage therapy involving single phages or phage cocktails, phage-derived enzymes, phages in combination with antibiotics, and genetically modified phages[2]. In this section we will summarize some of the main applications of phages and their by-products for the removal of biofilms (Figure 1).
Figure 1. Main phage-based treatments for biofilm removal.
2. Phage Therapy
Since phages can actively penetrate and disturb biofilms in nature, they can be used to obtain specific and improved treatments against biofilms[3]. Phage-based therapies focus on lytic phages because they destroy their bacterial hosts, but also because they lack integrases and other enzymes involved in horizontal gene transfer[4]. In order to design phage-based methods to remove biofilms, it is important to take into account the specific characteristics of the phages that may play a role in their penetration, diffusion, and propagation through the biofilm. For example, penetration of the biofilm is often less efficient for larger phages[5].
Phages encoding EPS-degrading enzymes are of particular interest against biofilms. Depolymerases are enzymes encoded by phages that specifically degrade EPS matrix components, improving phage penetration[6]. Another source of EPS-degrading enzymes are the bacteria found inside the biofilm under stress conditions. Stress can be triggered by phage infection, facilitating increased penetration and dissemination of phages within the community. This has been demonstrated in Pseudomonas aeruginosa biofilms, where phage infection was found to reduce the viscosity of biofilms by bacterial enzymes[7]. Phage therapy against biofilms of P. aeruginosa has been also tested in a mouse model of cystic fibrosis and has been shown to successfully remove biofilms[8]. Phages have also been shown to be effective against oral biofilms that cause infections such as caries, periodontal and peri-implant disease, including Enterococcus faecalis, Fusobacterium nucleatum, and Streptococcus spp. among others, suggesting promising new oral health products based on phages[9].
Antibiotics are usually broad-spectrum stable chemical compounds, while phages are very specific and evolving entities. On the one hand, their specificity is an advantage, as it reduces off-target damage and restricts the development of resistance to target-specific bacteria[10]. In addition, phages are evolving entities that can counteract bacterial resistance. On the other hand, specificity is also a limitation because it requires great efforts in terms of phage bioprospecting. Furthermore, specificity means that the bacterial pathogen has to be identified at species or even strain level before treatment is administered, which can be a problem for acute infections requiring a rapid response. This issue can be addressed by phage cocktails. If the target is a single species or strain, phages that do not infect the target will simply function as a bystander. However, biofilms are often multi-species communities, which means that cocktails can contribute to disrupting biofilms more efficiently[11]. Another interesting aspect of phage cocktails is that they can prevent the emergence of phage resistant bacteria if multiple phages active against a given target are included in the cocktail[12][13]. In addition, the phages within a cocktail can interact synergistically, increasing lytic activity[14]. However, interference or antagonistic interactions between phages could be also possible.
Recent studies support the use of cocktails against bacterial biofilms in vivo, especially for multi-species biofilms. For example, a phage cocktail was formulated to treat catheter-associated urinary tract infections caused by Proteus mirabilis, showing strong biofilm destruction activity and preventing biofilm formation. The application of this cocktail in liquid or gel form to rinse the urological catheters was proposed to cover their surface during application to prevent the formation of biofilms[15]. In addition, infections caused by P. aeruginosa biofilms were treated with a cocktail containing six lytic phages and tested with encouraging results[16]. Another cocktail combining six phages was tested to eradicate P. aeruginosa biofilms in a mice model with acute respiratory infection, showing great efficacy in disrupting biofilms[17]. Finally, two-phage cocktails are sometimes sufficient, as demonstrated to treat E. faecalis biofilms[18], although it is recommended to include more phages to reduce emergence of resistance.
Some phage-based products already on the market have been proposed as promising tools to remove biofilms[19], such a staphylococcal bacteriophage, containing the monophage Sb-1, which has been used in patients with osteomielytis and in foot ulcers[20], and PYO bacteriophage, a complex preparation designed for wound treatment[21] [64]. There are also some commercially available phage-based products for the food industry against Listeria sp.[22][23] or E. coli[24] with bactericidal effects that are interesting for biofilm prevention. Some of these commercially available products, as Listex P100[22] may also be promising for biofilm removal in surfaces of working environments of the food industry[25].
3. Phage-Derived Enzymes
Some enzymes encoded with phages may be useful for treating bacterial infections and biofilms[26]. These enzymes or enzybiotics derived from phages can be used as an alternative to antibiotics for human and animal health. Their efficacy has been demonstrated in a few pre-clinical studies, but these products are still under development. Under current safety standards and regulations, the application of phage products is easier than use of the phage itself. According to this view, two main types of phage degradation enzymes are useful in the removal of biofilms: lysins and depolymerases (Figure 2).
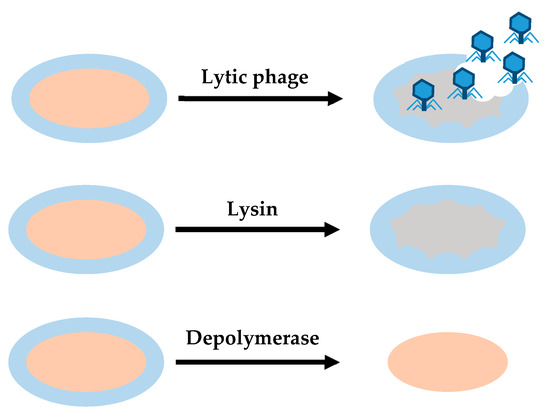
Figure 2. Differences between the action of lytic phages, lysins and depolymerases. Lytic phages provide antibacterial effect, degrading cell wall and EPS. Lysins provide a bactericidal effect, disrupting cell walls when they establish contact with their target. Depolymerases degrade EPS.
3.1. Lysins
Lysins are peptidoglycan hydrolases that have a bactericidal effect on susceptible bacteria. They break peptidoglycan bonds, degrading the bacterial cell wall and biofilm structure[27][28][29]. This makes lysins useful for Gram-positive bacteria[30]. Lysins are not restricted to be encoded by phages, since some bacteria produce lysins used to compete with other bacteria. In phages, lysins can be soluble enzymes, such as proteins that act at the end of the phage cycle to lysate the cell. In addition, they can be found in phage tails as virion-associated lysins, acting after receptor recognition to degrade the cell wall locally and allow injection of phage genomic material[31]. Depending on the peptidoglycan bonds they break, lysins are classified into different categories. Glycosidases or glycoside hydrolases break glycosidic bonds in complex sugars, N-acetylmuramoyl-L-alanine amidases cleaves the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides, and endopeptidases are proteolytic peptidases that break peptide bonds in non-terminal amino acids[32].
Phages that encode lysins have co-evolved with bacteria, so the binding domain of these enzymes evolved to target a unique and essential molecule in the cell wall, peptidoglycan, a well-preserved structure[28][33][34]. Lysins have been shown to exhibit thermostability, high ionic tolerance, and synergistic activity with antibiotics and other lysins[31]. In addition, lysins can be engineered to modify their target specificity and improve killing activity[35]. An example is the chimeric lysin Csl2, obtained by fusion of the catalytic domain of Cp1-7 lysozyme to the CW-7 repeats of the LySMP lysine from a Staphylococcus suis phage. It was designed to remove S. suis biofilms with positive results in vitro, and validated in vivo with a zebrafish infection model[36].
One of the main interesting features of lysins as therapeutic agents is that their activity is independent of the bacterial physiological state[36]. It was shown that the use of Art-175 lysine against multi-drug-resistant P. aeruginosa biofilms caused osmotic lysis independent of bacterial metabolism. This is relevant for biofilm removal because lysins can destroy persistent bacteria within biofilms, even at low metabolic rates[34].
3.2. Depolymerases
Depolymerases are enzymes derived from phages that facilitate the early stages of phage infection by degrading the extracellular substances of encapsulated bacteria, and may also help to reach phage receptors[37]. They are capable of degrading the chains of capsular polysaccharides, exopolysaccharides, and O-polysaccharides from lipopolysaccharides and peptidoglycan. All these substances may constitute the capsule of some free-living bacteria, but most of them are important components of the biofilm matrix. Depolymerases can be associated with virions, forming part of the phage particle, or be in soluble form. The latter type of depolymerase can be released during lysis of the bacterial cell[31]. Due to the ability of phage-encoded depolymerases to degrade the polysaccharides in the bacterial capsule and biofilm matrix, phages encoding these enzymes may have easier access to the bacterial host, allowing infection. Therefore, depolymerase activity is particularly interesting in the removal of biofilms, as it alters the EPS matrix and decreases bacterial virulence[37].
Depolymerases are divided into different groups. Hydrolases are depolymerases that use one molecule of water to hydrolyze chemical bonds, while lyases catalyze the breaking of chemical bonds by means other than hydrolysis and oxidation. A third type of depolymerases are triacylglycerol lipases. They act on the carboxylic ester bonds of triacylglycerols by releasing organic acids and glycerol. In addition to the diversity of depolymerase general modes of action, within each category there is also a great diversity and depolymerases are highly target-specific. This diversity and specificity is a result of phage-host co-evolution, influenced by intense horizontal gene transfer[31][37]. Depolymerases are especially interesting for treating human or animal infections caused by biofilms. They can enhance the action of the immune system against bacteria by degrading the EPS matrix and allowing immune cells to access the bacteria in the biofilm[26].
Depolymerases have been tested against biofilms formed by different bacterial species. Depolymerase Dpo7, derived from the vB_SepiS-phiPLA7 phage, was shown to reduce Staphylococcus sp. biofilm biomass by 53%–85% in 67% of the bacterial strains tested, in a dose-dependent but time-independent response[38]. Another example of depolymerase tested on biofilms with interesting results is Dpo42, derived from phage vB_EcoM_ECOO78. Its anti-biofilm activity was tested against Escherichia coli, again exhibiting dose-dependent biofilm prevention activity[39]. Finally, lysins and depolymerases are also good anti-biofilm agents in combination. For instance, lysin LysK and depolymerase DA7 have been tested in combination against Staphylococcus aureus biofilms in static and dynamic models. These enzymes showed a synergistic behavior, significantly reducing the number of viable cells in the biofilm[40].
4. Genetically Modified Phages
Penetration and diffusion of phages through the EPS-matrix is mandatory to eliminate biofilms using phage-based treatments. As mentioned above, some phage degradation enzymes serve this purpose, but many phages do not encode for these specific enzymes. However, phages can be genetically modified to produce enzymes that degrade the EPS-matrix, facilitating the removal of biofilms[41]. For example, a modified T7 E. coli phage has been designed to express intracellularly a hydrolase that is released during infection to the extracellular matrix, enhancing biofilm degradation. Testing on E. coli biofilms showed an elimination rate greater than 99%, and demonstrated the benefits of using manipulated phages[42].
Some temperate phages may have phenotypic characteristics that make them useful for biofilm removal. Genetic engineering can be used to turn these phages into lytic phages. This has been done by modifying the lysogenic ɸEf11 E. faecalis phage. E. faecalis biofilms are commonly associated with cases of failed root canals and nosocomial infections. ɸEf11 was genetically modified to eliminate all genes related to lysogeny, eliminating transduction problems and achieving a significant reduction in the biomass of treated E. faecalis biofilms, both resistant and sensitive to the antibiotic vancomycin[4].
Another interesting feature of genetically modified phages is related to host range. In a recent study, researchers modified the genome of T7Select E. coli phage by inserting coding sequences for 1080, a short peptide with a broad-spectrum anti-biofilm effect. The modified phage was more effective in eradicating established E. coli biofilms than the unmodified phage[43]. Phages can also be designed to selectively kill antibiotic resistant bacteria. In addition, although lytic phages are typically used to destroy bacteria, temperate phages may be of interest for delivering programmable DNA nucleases associated with CRISPR to reverse antibiotic resistance. This system can selectively destroy plasmids that confer antibiotic resistance[44].
Lytic phages infect host cells in order to replicate and release new virions, leading to an exponential increase of viral populations along time. In addition, as replicating evolving entities, phages could potentially induce gene transduction and other drawbacks. In order to avoid these problems, phages could be modified. An interesting example is phage AuNR, genetically modified to express a receptor-specific binding protein to attach to several Gram-negative organisms. In addition, they were conjugated to gold nanorods, that following excitation by near-infrared light, induced the photothermal lysis of the targeted cells, also destroying the phages and avoiding replication. This phage treatment was tested over P. aeruginosa biofilms, showing widespread bacterial cell death even when they were cultured in mammalian epithelial cells. Thus, combination of gold nanorods and genetically modified phages results in an interesting tool for biofilm removal[45].
5. Phages in Combination with Antibiotics
A sub-lethal dose of antibiotics can stimulate phage virulence under certain conditions. This phenomenon is known as phage-antibiotic synergy (PAS). The idea of combining phage therapy and antibiotics comes from the understanding that by using two different selective pressures we can obtain more efficacy than by using each separately[10][46]. An example of the success of the combination of phages and antibiotics was demonstrated in a study in which the Sb-1 S. aureus phage increased antibiotic activity against biofilms. Phage Sb-1 is particularly interesting for the treatment of S. aureus biofilms because of its ability to degrade the EPS-matrix[47]. Combination therapy of phages and antibiotics on E. coli biofilms has also been tested using T4 phages and tobramycin, which strongly reduced antibiotic-resistant bacteria. The same test was done for P. aeruginosa biofilms, using phage PB-1[48].
The combination of phage-derived enzymes with antibiotics, such as the combination of depolymerases with antibiotics, can increase the antibacterial effect by facilitating the access of antibiotics to the bacteria within the biofilm[26]. In a study of bacterial biofilms in food processing environments, the action of a thermally stable depolymerase obtained from a Klebsiella phage was tested. The enzymatic pre-treatment increased the subsequent disinfection effect of chlorine dioxide, a broad-spectrum sterilizer commonly used in the food industry. This enzyme reduced the adhesion of bacteria and EPS-matrix, favoring the action of chlorine dioxide[49].
However, it is important to note that the combination of phages and antibiotics also has some drawbacks. This may lead to the emergence of double-resistant bacteria, similar to antibiotic cocktails[50]. In addition, phages may preferentially infect antibiotic-sensitive bacteria compared to those that form antibiotic-resistant biofilms, further promoting antibiotic resistance[34][50]. Moreover, antibiotics could potentially interfere with bacterial metabolism, which is required for phages to infect bacteria. For these reasons, the effects of double treatment of phages with antibiotics should be tested to avoid incompatibilities, as antagonistic effects could arise[51][52][53].
Designing phage cocktails that include antibiotics has also been considered. The use of phage cocktails and antibiotics is especially interesting for treating multiple bacterial infections because some pathogenic species or strains may be favored by eliminating competitors[54].
References
- Pauline D. Scanlan; Angus Buckling; Alex R. Hall; Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research. Bacteriophage 2015, 5, e1050153, 10.1080/21597081.2015.1050153.
- Diana P. Pires; Luís D. R. Melo; Diana Vilas Boas; Miguel A. Cerqueira; Joana Azeredo; Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Current Opinion in Microbiology 2017, 39, 48-56, 10.1016/j.mib.2017.09.004.
- Pilar Domingo-Calap; Jennifer Delgado-Martínez; Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics 2018, 7, 66, 10.3390/antibiotics7030066.
- Justine Monnerat Tinoco; Bettina Buttaro; Hongming Zhang; Nadia Liss; Luciana M Sassone; Roy H. Stevens; Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms.. Archives of Oral Biology 2016, 71, 80-86, 10.1016/j.archoralbio.2016.07.001.
- Jun Hu; Kazuhiko Miyanaga; Yasunori Tanji; Diffusion properties of bacteriophages through agarose gel membrane. Biotechnology Progress 2010, 26, 1213-1221, 10.1002/btpr.447.
- Anneleen Cornelissen; Pieter-Jan Ceyssens; Jeroen T'syen; Helena Van Praet; Jean-Paul Noben; Olga V. Shaburova; Victor N. Krylov; Guido Volckaert; Rob Lavigne; The T7-Related Pseudomonas putida Phage ϕ15 Displays Virion-Associated Biofilm Degradation Properties. PLOS ONE 2011, 6, e18597, 10.1371/journal.pone.0018597.
- Geoffrey W. Hanlon; Stephen P. Denyer; Cedric J. Olliff; Lamia J. Ibrahim; Reduction in Exopolysaccharide Viscosity as an Aid to Bacteriophage Penetration through Pseudomonas aeruginosa Biofilms. Applied and Environmental Microbiology 2001, 67, 2746-2753, 10.1128/aem.67.6.2746-2753.2001.
- Elaine Waters; Daniel R Neill; Basak Kaman; Jaspreet S Sahota; Martha R J Clokie; Craig Winstanley; Aras Kadioglu; Phage therapy is highly effective against chronic lung infections withPseudomonas aeruginosa. Thorax 2017, 72, 666-667, 10.1136/thoraxjnl-2016-209265.
- Szymon Szafranski; Andreas Winkel; Meike Stiesch; The use of bacteriophages to biocontrol oral biofilms. Journal of Biotechnology 2017, 250, 29-44, 10.1016/j.jbiotec.2017.01.002.
- Amir Mohaghegh Motlagh; Ananda Bhattacharjee; Ramesh Goel; Biofilm control with natural and genetically-modified phages. World Journal of Microbiology and Biotechnology 2016, 32, 67, 10.1007/s11274-016-2009-4.
- Benjamin K Chan; Stephen Abedon; Catherine Loc-Carrillo; Phage cocktails and the future of phage therapy. Future Microbiology 2013, 8, 769-783, 10.2217/fmb.13.47.
- Anni-Maria Örmälä; Matti Jalasvuori; Phage therapy. Bacteriophage 2013, 3, e24219, 10.4161/bact.24219.
- Benjamin K. Chan; Stephen Abedon; Phage Therapy Pharmacology. Natural and Engineered Resistance to Plant Viruses, Part II 2012, 78, 1-23, 10.1016/b978-0-12-394805-2.00001-4.
- Matthew Schmerer; Ian J. Molineux; James J. Bull; Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590, 10.7717/peerj.590.
- Agnieszka Maszewska; Marta Zygmunt; Iwona Grzejdziak; Antoni Rózalski; Use of polyvalent bacteriophages to combat biofilm of Proteus mirabilis causing catheter-associated urinary tract infections. Journal of Applied Microbiology 2018, 125, 1253-1265, 10.1111/jam.14026.
- Diana R. Alves; Patricia Perez Esteban; Witold Kot; J.E. Bean; Tom Arnot; L.H. Hansen; Mark Enright; A. Toby A. Jenkins; A novel bacteriophage cocktail reduces and disperses P seudomonas aeruginosa biofilms under static and flow conditions. Microbial Biotechnology 2015, 9, 61-74, 10.1111/1751-7915.12316.
- Francesca Forti; Dwayne R. Roach; Marco Cafora; Maria E. Pasini; David S. Horner; Ersilia V. Fiscarelli; Martina Rossitto; Lisa Cariani; Federica Briani; Laurent Debarbieux; et al.Daniela Erica Ghisotti Design of a Broad-Range Bacteriophage Cocktail That Reduces Pseudomonas aeruginosa Biofilms and Treats Acute Infections in Two Animal Models. Antimicrobial Agents and Chemotherapy 2018, 62, e02573-17, 10.1128/aac.02573-17.
- Leron Khalifa; Daniel Gelman; Mor Shlezinger; Axel Lionel Dessal; Shunit Coppenhagen-Glazer; Nurit Beyth; Ronen Hazan; Defeating Antibiotic- and Phage-Resistant Enterococcus faecalis Using a Phage Cocktail in Vitro and in a Clot Model. Frontiers in Microbiology 2018, 9, 326, 10.3389/fmicb.2018.00326.
- Tamta Tkhilaishvili; Lei Wang; Arianna Tavanti; Andrej Trampuz; Mariagrazia Di Luca; Antibacterial Efficacy of Two Commercially Available Bacteriophage Formulations, Staphylococcal Bacteriophage and PYO Bacteriophage, Against Methicillin-Resistant Staphylococcus aureus: Prevention and Eradication of Biofilm Formation and Control of a Systemic Infection of Galleria mellonella Larvae.. Frontiers in Microbiology 2020, 11, 110, 10.3389/fmicb.2020.00110.
- R. Fish; E. Kutter; G. Wheat; B. Blasdel; M. Kutateladze; Sarah Kuhl; Bacteriophage treatment of intransigent diabetic toe ulcers: a case series. Journal of Wound Care 2016, 25, 27–33, 10.12968/jowc.2016.25.7.s27.
- Kalistrat Markoishvili; George Tsitlanadze; Ramaz Katsarava; J Glenn Morris; Alexander Sulakvelidze; A novel sustained-release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds.. International Journal of Dermatology 2002, 41, 453–458.
- PhageGuard Listeria. Available online: https://phageguard.com/es/solucion-listeria/ (accessed on 14 May 2020)
- Intralytix, Inc. Available online: http://www.intralytix.com/index.php?page=prod&id=1 (accessed on 14 May 2020)
- Intralytix, Inc. Available online: http://www.intralytix.com/index.php?page=prod&id=2 (accessed on 14 May 2020)
- Lucilla Iacumin; Marisa Manzano; Giuseppe Comi; Phage Inactivation of Listeria monocytogenes on San Daniele Dry-Cured Ham and Elimination of Biofilms from Equipment and Working Environments. Microorganisms 2016, 4, 4, 10.3390/microorganisms4010004.
- Barbara Maciejewska; Tomasz Olszak; Zuzanna Drulis-Kawa; Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application?. Applied Microbiology and Biotechnology 2018, 102, 2563-2581, 10.1007/s00253-018-8811-1.
- Roberto Vázquez; Ernesto García; Pedro García; Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Frontiers in Immunology 2018, 9, 2252, 10.3389/fimmu.2018.02252.
- Daniel B. Gilmer; Jonathan E. Schmitz; Chad W. Euler; Vincent A. Fischetti; Novel Bacteriophage Lysin with Broad Lytic Activity Protects against Mixed Infection by Streptococcus pyogenes and Methicillin-Resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 2013, 57, 2743-2750, 10.1128/aac.02526-12.
- Umender Sharma; Aradhana Vipra; Shankaramurthy Channabasappa; Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discovery Today 2018, 23, 848-856, 10.1016/j.drudis.2018.01.026.
- Franklin L. Nobrega; Ana Rita Costa; L. D. Kluskens; Joana Azeredo; Revisiting phage therapy: new applications for old resources. Trends in Microbiology 2015, 23, 185-191, 10.1016/j.tim.2015.01.006.
- Agnieszka Latka; Barbara Maciejewska; Grazyna Majkowska-Skrobek; Yves Briers; Zuzanna Drulis-Kawa; Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Applied Microbiology and Biotechnology 2017, 101, 3103-3119, 10.1007/s00253-017-8224-6.
- Daniel C. Nelson; Lawrence Loomis; Vincent A. Fischetti; Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proceedings of the National Academy of Sciences 2001, 98, 4107-4112, 10.1073/pnas.061038398.
- Juan A. Hermoso; Ernesto García; Pedro García; Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Current Opinion in Microbiology 2007, 10, 461-472, 10.1016/j.mib.2007.08.002.
- Yves Briers; Maarten Walmagh; Barbara Grymonprez; Manfred Biebl; Jean-Paul Pirnay; Valerie DeFraine; Jan Michiels; William Cenens; Abram Aertsen; Stefan Miller; et al.Rob Lavigne Art-175 Is a Highly Efficient Antibacterial against Multidrug-Resistant Strains and Persisters of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy 2014, 58, 3774-3784, 10.1128/aac.02668-14.
- Rubens López; Ernesto García; Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiology Reviews 2004, 28, 553-580, 10.1016/j.femsre.2004.05.002.
- Roberto Vázquez; Mirian Domenech; Manuel Iglesias-Bexiga; Margarita Menendez; Pedro García; Csl2, a novel chimeric bacteriophage lysin to fight infections caused by Streptococcus suis, an emerging zoonotic pathogen.. Scientific Reports 2017, 7, 16506, 10.1038/s41598-017-16736-0.
- Diana P. Pires; Hugo Oliveira; Luís D. R. Melo; Miguel A. Cerqueira; Joana Azeredo; Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Applied Microbiology and Biotechnology 2016, 100, 2141-2151, 10.1007/s00253-015-7247-0.
- Diana Gutiérrez; Yves Briers; Lorena Rodriguez-Rubio; Beatriz Martínez; Ana Rodríguez; Rob Lavigne; Pilar García; Role of the Pre-neck Appendage Protein (Dpo7) from Phage vB_SepiS-phiIPLA7 as an Anti-biofilm Agent in Staphylococcal Species. Frontiers in Microbiology 2015, 6, 1315, 10.3389/fmicb.2015.01315.
- Zhimin Guo; Jing Huang; Guangmou Yan; Liancheng Lei; Shuang Wang; Ling Yu; Liang Zhou; Anchong Gao; Xin Feng; Wenyu Han; et al.Jingmin GuJunling Yang Identification and Characterization of Dpo42, a Novel Depolymerase Derived from the Escherichia coli Phage vB_EcoM_ECOO78. Frontiers in Microbiology 2017, 8, 1460, 10.3389/fmicb.2017.01460.
- Nanna M. C. Olsen; Elowine Thiran; Tobias Hasler; Thomas Vanzieleghem; Georgios N. Belibasakis; Jacques Mahillon; Martin J. Loessner; Mathias Schmelcher; Synergistic Removal of Static and Dynamic Staphylococcus aureus Biofilms by Combined Treatment with a Bacteriophage Endolysin and a Polysaccharide Depolymerase. Viruses 2018, 10, 438, 10.3390/v10080438.
- Rodney M. Donlan; Preventing biofilms of clinically relevant organisms using bacteriophage. Trends in Microbiology 2009, 17, 66-72, 10.1016/j.tim.2008.11.002.
- Timothy K. Lu; James J. Collins; Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences 2007, 104, 11197-11202, 10.1073/pnas.0704624104.
- David J. Lemon; Matthew K. Kay; James K. Titus; April A. Ford; Wen Chen; Nicholas J. Hamlin; Yoon Y. Hwang; Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. The Journal of Microbiology 2019, 57, 532-538, 10.1007/s12275-019-8686-6.
- Ido Yosef; Miriam Manor; Ruth Kiro; Udi Qimron; Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proceedings of the National Academy of Sciences 2015, 112, 7267-7272, 10.1073/pnas.1500107112.
- Huan Peng; Raymond E. Borg; Liam P. Dow; Beth L. Pruitt; Irene A. Chen; Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proceedings of the National Academy of Sciences 2020, 117, 1951-1961, 10.1073/pnas.1913234117.
- Clara Torres-Barceló; Michael E. Hochberg; Evolutionary Rationale for Phages as Complements of Antibiotics. Trends in Microbiology 2016, 24, 249-256, 10.1016/j.tim.2015.12.011.
- Tamta Tkhilaishvili; Lisa Lombardi; Ann-Brit Klatt; Andrej Trampuz; Mariagrazia Di Luca; Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. International Journal of Antimicrobial Agents 2018, 52, 842-853, 10.1016/j.ijantimicag.2018.09.006.
- Lindsey B. Coulter; Robert J.C. McLean; Rodney E. Rohde; Gary M. Aron; Effect of Bacteriophage Infection in Combination with Tobramycin on the Emergence of Resistance in Escherichia coli and Pseudomonas aeruginosa Biofilms. Viruses 2014, 6, 3778-3786, 10.3390/v6103778.
- Zihan Chai; Jing Wang; Suyuan Tao; Haijin Mou; Application of bacteriophage-borne enzyme combined with chlorine dioxide on controlling bacterial biofilm. LWT 2014, 59, 1159-1165, 10.1016/j.lwt.2014.06.033.
- Rafael Peña-Miller; David Lähnemann; Gunther Jansen; Ayari Fuentes-Hernandez; Philip Rosenstiel; Hinrich Schulenburg; Robert E Beardmore; When the Most Potent Combination of Antibiotics Selects for the Greatest Bacterial Load: The Smile-Frown Transition. PLOS Biology 2013, 11, e1001540, 10.1371/journal.pbio.1001540.
- Stephen T. Abedon; Phage-Antibiotic Combination Treatments: Antagonistic Impacts of Antibiotics on the Pharmacodynamics of Phage Therapy?. Antibiotics 2019, 8, 182, 10.3390/antibiotics8040182.
- Thaysa Tagliaferri; Mathias Jansen; Hans-Peter Horz; Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy.. Frontiers in Microbiology 2019, 9, 22, 10.3389/fcimb.2019.00022.
- Takeru Matsui; Genki Yoshikawa; Tomoko Mihara; Orawan Chatchawankanphanich; Takeru Kawasaki; Miyako Nakano; Makoto Fujie; Hiroyuki Ogata; Takashi Yamada; Replications of Two Closely Related Groups of Jumbo Phages Show Different Level of Dependence on Host-encoded RNA Polymerase. Frontiers in Microbiology 2017, 8, 1010, 10.3389/fmicb.2017.01010.
- Susan M. Lehman; Rodney M. Donlan; Bacteriophage-Mediated Control of a Two-Species Biofilm Formed by Microorganisms Causing Catheter-Associated Urinary Tract Infections in anIn VitroUrinary Catheter Model. Antimicrobial Agents and Chemotherapy 2014, 59, 1127-1137, 10.1128/aac.03786-14.




