
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rui Vitorino | + 3827 word(s) | 3827 | 2021-05-07 11:07:28 | | | |
| 2 | Dean Liu | -10 word(s) | 3817 | 2021-06-21 11:32:02 | | | | |
| 3 | Dorian Hanaor | + 16 word(s) | 3833 | 2024-03-05 18:42:13 | | |
Video Upload Options
Microfluidics is the advanced microtechnology of fluid manipulation in channels with at least one dimension in the range of 1–100 microns. Microfluidic technology offers a growing number of tools for manipulating small volumes of fluid to control chemical, biological, and physical processes relevant to separation, analysis, and detection.
1. Introduction
Microfluidics (MF) is a relatively new branch of science and microengineering that deals with manipulating fluids in microchannels with at least one dimension of 1 to 100 micrometers [1][2], as shown in Figure 1. Scientists consider it a new discipline not only because of the recent emergence of microfluidic (MF) devices that can implement rapid solutions to complex analytical problems at the microscale but also because the physical principles of fluid flow at such small length scales differ from those in macrosystems [3]. MF spans several disciplines—physical and chemical sciences, micromechanics, electronics, and mechanical engineering. It has wide applicability in many fields, with particular emphasis on biology, biochemistry, biotechnology, medicine, pharmacology, and food and environmental analysis. It has made great progress in the last 15 to 20 years [4]. Intensively developing research areas in MF are lab-on-chip (LOC) devices [5][6] and microanalytical systems (μTAS) [7]. They can be considered synonymous with integrated circuits in electronics. The MF technology is based on micropumps, mixers, filters and valves to realize chemical and biological laboratory processes on a single chip. MF requires tiny samples and reagents for analysis, which makes it environmentally friendly and minimally invasive [8].
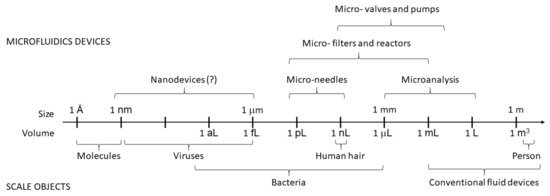
Figure 1. Scales and volumes in microfluidics.
MF is perceived as a new platform for highly efficient separation and highly sensitive analysis of (bio)molecules and (bio)particles in biochemistry, molecular biology and biotechnology [8] and relies on scale reduction to reduce material consumption and cost [1]. MF exploits the potential of flowing liquids at the microscale to generate “quantitative assays”. In peptidomics and proteomics, MF is most commonly used in combination with MS analyses. Hydrophobic membranes were originally used to adsorb native peptides or peptides generated by enzymatic digestion of proteins, followed by their desalting and elution in a controlled environment for MS analysis [1][2]. Peptides and proteins are analyzed at MS mainly by two ionization techniques, electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI). ESI–MS and MALDI–MS are used not only to determine the relative molecular masses of peptides and proteins but also to elucidate their amino acid sequences and post-translational modifications [9][10][11][12]). Rapid and highly sensitive analysis was achieved by using ultralow sample volumes and amounts (picomoles to femtomoles of compounds in nanoliters to picoliters of volume per single analysis), high separation efficiency, and short analysis times. The widespread use of MF can be attributed to the inherent advantages of MF instruments: Wide applicability, compactness and the need for extremely low sample volumes of the analyzed compounds or particles, as well as low reagent consumption [2][12][13]. By tailoring the geometry of pores in porous materials, fluid flow can be controlled in microfluidic devices. [14]
2. Applications of Microfluidics
3.1. Analysis of Peptides and Proteins; Peptidomics and Proteomics
Peptides and proteins are extremely important biological molecules. As hormones, hormone or drug receptors, enzymes, coenzymes, enzyme substrates or inhibitors, antigens, antibodies, immunomodulators, antibiotics, structural elements and transport molecules play a crucial role in all living organisms. They ensure the basic operations of the cellular machinery. In addition, there are many peptide and protein-based drugs and prodrugs, and some peptides and proteins are used as biomarkers [15]. Moreover, a complete analysis of all peptides (peptidomes) and proteins (proteomes) of a cell, tissue, biofluid, organ or organism is important to understand normal and pathological processes. This is the subject of peptidomics and proteomics—comprehensive and large-scale studies of complex mixtures of peptides and proteins. In this context, the relevance of peptides is increasing because the structure and function of proteins are often identified by their enzymatically generated peptide fragments. This bottom-up or shot-gun approach is one of the main directions in current proteomics research.
The separation and study of peptides and proteins in complex biological matrices is a challenging process that requires advanced and accurate methods that can provide relevant information about their structural and functional properties.
It has been known for more than a century that proteins can move or even “fly” under the influence of an electric field [16]. Therefore, electromigration methods represent powerful tools for their separation and analysis. Polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS–PAGE), isoelectric gel focusing (IEF), and two-dimensional gel electrophoresis (2-DE), which combines the orthogonal principles of narrow tube gel IEF in the first dimension and plate gel SDS–PAGE in the second dimension, have been widely used in the past for protein and polypeptide analysis [17][18]. Currently, HPLC and UHPLC combined with high-resolution MS detection are the leading techniques for peptide and protein analysis [19]. Capillary and microchip electromigration methods (CE/MCE) are also very powerful and useful methods for the analysis and characterization of peptides and proteins [20][21]) and for applications in peptidomics and proteomics [22]. CE and MCE methods have several advantages, such as high separation efficiency, short analysis time, low sample and reagent consumption, and different separation modes (ZE, ITP, IEF, AE, EKC, and EC). In the field of peptidomics and proteomics, CE and MCE are usually combined with MS detection, as MS can identify and quantify the separated peptides and proteins. In addition to MS detection, UV absorption or LIF detection are also commonly used in CE and MCE analyses of peptides and proteins [21][23].
Biological samples (e.g., body fluids, tissues, and food extracts) contain complex mixtures of variable compounds with low and high molecular mass. Therefore, the target peptides and proteins usually need to be extracted or pre-isolated and/or preconcentrated from the sample matrix before analysis [24][25]. The method of sample preconcentration is based on (i) the principle of electrophoresis [26][27][28] (on-site field-assisted sample stacking, micelle stacking, ITP and IEF) or (ii) selective adsorption/extraction method (elution) [29][30]. Usually, a combination of different types of preconcentrates is used. Another important issue in CE and MCE analyses of peptides and proteins is the prevention of protein adsorption on the inner wall of the capillary.
CE/MCE technology was introduced to improve the efficiency of biomarker protein analysis. This technology can find wide applications in hospitals and other immediate medical facilities. This has been proved by several studies of protein processing related to biomedical research and application. Štěpánová and Kašička [21] and Dawod et al. [6][31] described the developments in protein analysis using various CE and MCE methods in 2011–2017. They showed that sample preparation, preconcentration, inhibition of adsorption and control of EOF, separation by a specific CE/MCE method and improvement of the detection scheme have greatly improved the ability of CE/MCE methods for protein analysis. The innovative application of CE and MCE methods in biopharmaceutical protein quality control, protein determination in complex biometrics, peptide-protein mapping, and determination of physical and chemical parameters of proteins are important achievements in this field.
However, a faster and more sensitive analysis than existing analytical methods is needed.
The application of MF technology in this field is an intensively developed concept to create integrated and fully automated analytical devices that can detect and quantify one or more peptides and proteins from a complex matrix. In this miniaturized MF form of CE, all operations (including sample preparation, derivatization, injection, separation, and detection) are integrated into a micrototal analytical system (uTAS) or lab on chip (LOC) platform [32].
Sonker et al. [33] reported the possibility of using a specific separation system for integrated immunoaffinity extraction to study human serum matrix biomarkers in preterm infants. They used a reactive polymer to immobilize the antibody as a whole and selectively extracted targeted preterm infant markers. For effective separation, they also optimized the MF immunoaffinity extraction protocol and combined it with MCE. The low nanomolar concentration of the two enriched preterm markers in the human serum matrix was studied for 30 min. Their observations may help to develop automated and integrated birth risk assessment tools.
Peptide identification by MS implicitly provides information about post-translational modification (PTM) sites. It can be used to identify the threshold of a particular enzyme under physiologically appropriate conditions. Noach-Hirsh et al. [34] presented a modular integrated MF platform to analyze multiple post-translational modifications of newly synthesized protein arrays. This method can also be used to clarify PTM fingerprints on single cells or tissues. Although the technology is comprehensive, it is limited in size and requires relatively small amounts of biological materials and reagents for research. It is suitable for basic and translational research [34].
More attention has been paid to tools used to identify target proteins or disease biomarkers. The most difficult challenge remains the ability to detect low protein abundances in a single cell. Recent advances in high-resolution/high-quality high-precision mass spectrometers have enabled identifying more than 5000 proteins from less than 100 ng of protein extracts in a short analysis time of 15 min LC–MS [35].
The crude protein content of mammalian cells is only about two orders of magnitude lower than the value that gives a detailed cell extract profile. There is also a need to improve the detection limit of MS, scan rate and intelligent data acquisition technology so that MF proteomics and single-cell or relatively small protein proteomics provide comparable results. Therefore, some researchers believe that MS detection strategies using existing data are most likely to achieve the required performance [35].
There are many important advances in the MF platform that can quantify unicellular proteins. Absolute quantification is the key criterion for the determination of unicellular proteins. Without absolute quantification, it is impossible to accurately compare protein amounts determined by different methods. Research to improve the precision of single-cell protein quantification also has great potential. Fluorescent NPs with higher fluorescence intensity can be used to replace conventional fluorescent probes. Nucleic acid-labeled antibodies can be further amplified and quantified in automated PCR [36].
3.2. Separation and Analysis of Cells
3.2.1. Cell Sorting and Single-Cell Analysis
3.2.2. Secretome Analysis and Single-Cell Omics Analysis
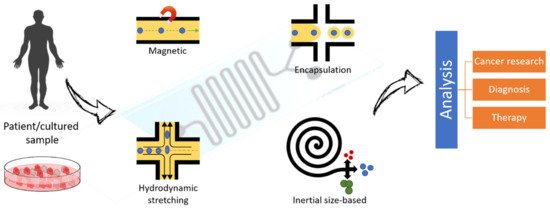
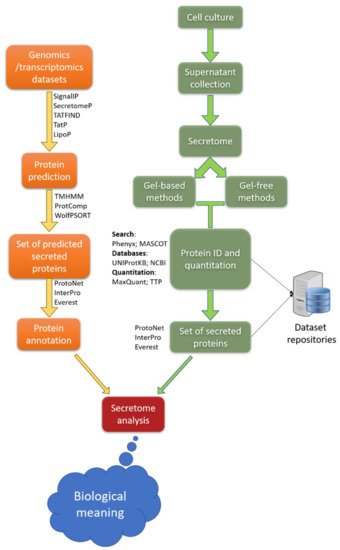
3.2.3. Investigation of Stimulus-Driven Cell Behavior
3.2.4. Investigation of Biomolecular Coronas Nanoparticles
References
- Lee, W.; Tseng, P.; Di Carlo, D. Microfluidic Cell Sorting and Separation Technology. In Microtechnology for Cell Manipulation and Sorting; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–14.
- Gao, D.; Song, C.; Lin, J.-M. Microfluidics-Mass Spectrometry Combination Systems for Single-Cell Analysis. In Microfluidics for Single-Cell Analysis; Springer: Berlin/Heidelberg, Germany, 2019; pp. 163–195.
- Liu, Y.; Yang, Q.; Cao, L.; Xu, F. Analysis of Leukocyte Behaviors on Microfluidic Chips. Adv. Healthc. Mater. 2019, 8, 1801406.
- Chiu, D.T.; deMello, A.J.; Di Carlo, D.; Doyle, P.S.; Hansen, C.; Maceiczyk, R.M.; Wootton, R.C. Small but perfectly formed? Successes, challenges, and opportunities for microfluidics in the chemical and biological sciences. Chem 2017, 2, 201–223.
- Conde, J.P.; Madaboosi, N.; Soares, R.R.; Fernandes, J.T.S.; Novo, P.; Moulas, G.; Chu, V. Lab-on-chip systems for integrated bioanalyses. Essays Biochem. 2016, 60, 121–131.
- Narayanamurthy, V.; Jeroish, Z.; Bhuvaneshwari, K.; Bayat, P.; Premkumar, R.; Samsuri, F.; Yusoff, M.M. Advances in passively driven microfluidics and lab-on-chip devices: A comprehensive literature review and patent analysis. RSC Adv. 2020, 10, 11652–11680.
- Manz, A.; Graber, N.; Widmer, H.Á. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248.
- Bragheri, F.; Vázquez, R.M.; Osellame, R. Microfluidics. In Three-Dimensional Microfabrication Using Two-Photon Polymerization; Elsevier: Amsterdam, The Netherlands, 2020; pp. 493–526.
- Bai, Y.; Gao, M.; Wen, L.; He, C.; Chen, Y.; Liu, C.; Fu, X.; Huang, S. Applications of Microfluidics in Quantitative Biology. Biotechnol. J. 2018, 13, e1700170.
- Loo, J.A.; Udseth, H.R.; Smith, R.D. Peptide and protein analysis by electrospray ionization-mass spectrometry and capillary electrophoresis-mass spectrometry. Anal. Biochem. 1989, 179, 404–412.
- Kammeijer, G.S.; Kohler, I.; Jansen, B.C.; Hensbergen, P.J.; Mayboroda, O.A.; Falck, D.; Wuhrer, M. Dopant enriched nitrogen gas combined with sheathless capillary electrophoresis–electrospray ionization-mass spectrometry for improved sensitivity and repeatability in glycopeptide analysis. Anal. Chem. 2016, 88, 5849–5856.
- Barry, R.; Ivanov, D. Microfluidics in biotechnology. J. Nanobiotechnol. 2004, 2, 2.
- Soloviev, M.; Barry, R.; Scrivener, E.; Terrett, J. Combinatorial peptidomics: A generic approach for protein expression profiling. J. Nanobiotechnol. 2003, 1, 4.
- Mingchao Liu; Si Suo; Jian Wu; Yixiang Gan; Dorian Ah Hanaor; C.Q. Chen; Tailoring porous media for controllable capillary flow. J. Colloid Interface Sci.. 2018, 539, 379-387.
- Wei, Q.; Becherer, T.; Angioletti-Uberti, S.; Dzubiella, J.; Wischke, C.; Neffe, A.T.; Lendlein, A.; Ballauff, M.; Haag, R. Protein interactions with polymer coatings and biomaterials. Angew. Chem. Int. Ed. 2014, 53, 8004–8031.
- Burger, R.; Amato, L.; Boisen, A. Detection methods for centrifugal microfluidic platforms. Biosens. Bioelectron. 2016, 76, 54–67.
- Srinivas, P.R. Introduction to Protein Electrophoresis. In Electrophoretic Separation of Proteins; Springer: Berlin/Heidelberg, Germany, 2019; pp. 23–29.
- Westermeier, R. Looking at proteins from two dimensions: A review on five decades of 2D electrophoresis. Arch. Physiol. Biochem. 2014, 120, 168–172.
- Righetti, P.G.; Candiano, G. Recent advances in electrophoretic techniques for the characterization of protein biomolecules: A poker of aces. J. Chromatogr. A 2011, 1218, 8727–8737.
- Fekete, S.; Guillarme, D.; Sandra, P.; Sandra, K. Chromatographic, electrophoretic, and mass spectrometric methods for the analytical characterization of protein biopharmaceuticals. Anal. Chem. 2016, 88, 480–507.
- Kašička, V. Recent developments in capillary and microchip electroseparations of peptides (2017–mid 2019). Electrophoresis 2020, 41, 10–35.
- Štěpánová, S.; Kašička, V. Recent applications of capillary electromigration methods to separation and analysis of proteins. Anal. Chim. Acta 2016, 933, 23–42.
- Štěpánová, S.; Kašička, V. Recent developments and applications of capillary and microchip electrophoresis in proteomics and peptidomics (2015–mid 2018). J. Sep. Sci. 2019, 42, 398–414.
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693.
- Puangpila, C.; Mayadunne, E.; El Rassi, Z. Liquid phase based separation systems for depletion, prefractionation, and enrichment of proteins in biological fluids and matrices for in-depth proteomics analysis—An update covering the period 2011–2014. Electrophoresis 2015, 36, 238–252.
- Giordano, B.C.; Burgi, D.S.; Hart, S.J.; Terray, A. On-line sample pre-concentration in microfluidic devices: A review. Anal. Chim. Acta 2012, 718, 11–24.
- Gharari, H.; Farjaminezhad, M.; Marefat, A.; Fakhari, A.R. All-in-one solid-phase microextraction: Development of a selective solid-phase microextraction fiber assembly for the simultaneous and efficient extraction of analytes with different polarities. J. Sep. Sci. 2016, 39, 1709–1716.
- Bang, Y.; Hwang, Y.; Lee, S.; Park, S.; Bae, S. Sol–gel-adsorbent-coated extraction needles to detect volatile compounds in spoiled fish. J. Sep. Sci. 2017, 40, 3839–3847.
- Dawod, M.; Arvin, N.E.; Kennedy, R.T. Recent advances in protein analysis by capillary and microchip electrophoresis. Analyst 2017, 142, 1847–1866.
- Ríos, Á.; Zougagh, M.; Avila, M. Miniaturization through lab-on-a-chip: Utopia or reality for routine laboratories? A review. Anal. Chim. Acta 2012, 740, 1–11.
- Sonker, M.; Parker, E.K.; Nielsen, A.V.; Sahore, V.; Woolley, A.T. Electrokinetically operated microfluidic devices for integrated immunoaffinity monolith extraction and electrophoretic separation of preterm birth biomarkers. Analyst 2018, 143, 224–231.
- Noach-Hirsh, M.; Nevenzal, H.; Glick, Y.; Chorni, E.; Avrahami, D.; Barbiro-Michaely, E.; Gerber, D.; Tzur, A. Integrated microfluidics for protein modification discovery. Mol. Cell. Proteom. 2015, 14, 2824–2832.
- Lazar, I.M.; Gulakowski, N.S.; Lazar, A.C. Protein and proteome measurements with microfluidic chips. Anal. Chem. 2019, 92, 169–182.
- Fan, B.; Li, X.; Chen, D.; Peng, H.; Wang, J.; Chen, J. Development of microfluidic systems enabling high-throughput single-cell protein characterization. Sensors 2016, 16, 232.
- Santos, R.S.; Figueiredo, C.; Azevedo, N.F.; Braeckmans, K.; De Smedt, S.C. Nanomaterials and molecular transporters to overcome the bacterial envelope barrier: Towards advanced delivery of antibiotics. Adv. Drug Deliv. Rev. 2018, 136, 28–48.
- Kolarević, S.; Milovanović, D.; Avdović, M.; Oalđe, M.; Kostić, J.; Sunjog, K.; Nikolić, B.; Knežević-Vukčević, J.; Vuković-Gačić, B. Optimisation of the microdilution method for detection of minimum inhibitory concentration values in selected bacteria. Bot. Serbica 2016, 40, 29–36.
- Sollier, E.; Kochersperger, M.L.; Englert, R.F.; Che, J.; Huang, K.-W.; Boyce-Jacino, M.; Neddersen, A.; Passernig, A.; Richardson, B.; Choi, I. Microfluidic Chips and Cartridges and Systems Utilizing Microfluidic Chips and Cartridges. International Patent Application No. PCT/US2017/027959, 17 April 2017.
- Wang, Y.; Li, Q.; Shi, H.; Tang, K.; Qiao, L.; Yu, G.; Ding, C.; Yu, S. Microfluidic Raman biochip detection of exosomes: A promising tool for prostate cancer diagnosis. Lab Chip 2020, 20, 4632–4637.
- Kang, J.H.; Park, J.-K. Technical paper on microfluidic devices-cell separation technology. Asia Pac. Biotech News 2005, 9, 1135–1146.
- Chiu, Y.-Y.; Huang, C.-K.; Lu, Y.-W. Enhancement of microfluidic particle separation using cross-flow filters with hydrodynamic focusing. Biomicrofluidics 2016, 10, 011906.
- Menon, N.V.; Lim, S.B.; Lim, C.T. Microfluidics for personalized drug screening of cancer. Curr. Opin. Pharmacol. 2019, 48, 155–161.
- Kulasinghe, A.; Wu, H.; Punyadeera, C.; Warkiani, M.E. The use of microfluidic technology for cancer applications and liquid biopsy. Micromachines 2018, 9, 397.
- Pakjesm Pourfard, P. Single Cell Enrichment with High Throughput Microfluidic Devices. Master’s Thesis, University of California Irvine, Irvine, CA, USA, 2017.
- Zhu, P.; Wang, L. Passive and active droplet generation with microfluidics: A review. Lab Chip 2017, 17, 34–75.
- Lei, C.; Kobayashi, H.; Wu, Y.; Li, M.; Isozaki, A.; Yasumoto, A.; Mikami, H.; Ito, T.; Nitta, N.; Sugimura, T. High-throughput imaging flow cytometry by optofluidic time-stretch microscopy. Nat. Protoc. 2018, 13, 1603–1631.
- Liu, N.; Petchakup, C.; Tay, H.M.; Li, K.H.H.; Hou, H.W. Spiral Inertial Microfluidics for Cell Separation and Biomedical Applications. In Applications of Microfluidic Systems in Biology and Medicine; Springer: Berlin/Heidelberg, Germany, 2019; pp. 99–150.
- Ericson, C.; Holm, J.; Ericson, T.; Hjerten, S. Electroosmosis-and pressure-driven chromatography in chips using continuous beds. Anal. Chem. 2000, 72, 81–87.
- Su, W.; Li, H.; Chen, W.; Qin, J. Microfluidic strategies for label-free exosomes isolation and analysis. TrAC Trends Anal. Chem. 2019.
- Chagas, C.L.; Moreira, R.C.; Bressan, L.P.; de Jesus, D.P.; da Silva, J.A.; Coltro, W.K. Instrumental Platforms for Capillary and Microchip Electromigration Separation Techniques. In Capillary Electromigration Separation Methods; Elsevier: Amsterdam, The Netherlands, 2018; pp. 269–292.
- Pamme, N. Continuous flow separations in microfluidic devices. Lab Chip 2007, 7, 1644–1659.
- Kielpinski, M.; Walther, O.; Cao, J.; Henkel, T.; Köhler, J.M.; Groß, G.A. Microfluidic Chamber Design for Controlled Droplet Expansion and Coalescence. Micromachines 2020, 11, 394.
- Wang, J.; Zhang, N.; Chen, J.; Rodgers, V.G.; Brisk, P.; Grover, W.H. Finding the optimal design of a passive microfluidic mixer. Lab Chip 2019, 19, 3618–3627.
- Prakash, J.; Tripathi, D. Electroosmotic flow of Williamson ionic nanoliquids in a tapered microfluidic channel in presence of thermal radiation and peristalsis. J. Mol. Liq. 2018, 256, 352–371.
- Ahmad, I.L.; Ahmad, M.R.; Takeuchi, M.; Nakajima, M.; Hasegawa, Y. Tapered Microfluidic for Continuous Micro-Object Separation Based on Hydrodynamic Principle. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 1413–1421.
- Ahmad, I.L.; Ahmad, M.R. Tapered microchannel for multi-particles passive separation based on hydrodynamic resistance. Indones. J. Electr. Eng. Comput. Sci. 2017, 5, 628–635.
- Voigt, A.; Schreiter, J.; Frank, P.; Pini, C.; Mayr, C.; Richter, A. Method for the Computer-aided Schematic Design and Simulation of Hydrogel-based Microfluidic Systems. IEEE Trans. Comput. Aided Des. Integr. Circuits Syst. 2019.
- Naderi, A.; Bhattacharjee, N.; Folch, A. Digital Manufacturing for Microfluidics. Annu. Rev. Biomed. Eng. 2019, 21, 325–364.
- Sun, J.; Moore, L.; Xue, W.; Kim, J.; Zborowski, M.; Chalmers, J.J. Correlation of simulation/finite element analysis to the separation of intrinsically magnetic spores and red blood cells using a microfluidic magnetic deposition system. Biotechnol. Bioeng. 2018, 115, 1288–1300.
- Shamloo, A.; Boodaghi, M. Design and simulation of a microfluidic device for acoustic cell separation. Ultrasonics 2018, 84, 234–243.
- Hu, Q.; Luni, C.; Elvassore, N. Microfluidics for secretome analysis under enhanced endogenous signaling. Biochem. Biophys. Res. Commun. 2018, 497, 480–484.
- Liu, F.; KC, P.; Ni, L.; Zhang, G.; Zhe, J. A microfluidic competitive immuno-aggregation assay for high sensitivity cell secretome detection. Organogenesis 2018, 14, 67–81.
- Xu, X.; Wang, J.; Wu, L.; Guo, J.; Song, Y.; Tian, T.; Wang, W.; Zhu, Z.; Yang, C. Microfluidic Single-Cell Omics Analysis. Small 2020, 16, 1903905.
- Caen, O.; Lu, H.; Nizard, P.; Taly, V. Microfluidics as a strategic player to decipher single-cell omics? Trends Biotechnol. 2017, 35, 713–727.
- Deng, Y.; Finck, A.; Fan, R. Single-cell omics analyses enabled by microchip technologies. Annu. Rev. Biomed. Eng. 2019, 21, 365–393.
- Lazar, I.M.; Deng, J.; Stremler, M.A.; Ahuja, S. Microfluidic reactors for advancing the MS analysis of fast biological responses. Microsyst. Nanoeng. 2019, 5, 1–16.
- Bahrami, S.; Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016, 62, 37–49.
- Fowler, T.; Sen, R.; Roy, A.L. Regulation of primary response genes. Mol. Cell 2011, 44, 348–360.
- Dungan, J.; Mathews, J.; Levin, M.; Koomson, V. Microfluidic platform to study intercellular connectivity through on-chip electrical impedance measurement. In Proceedings of the 2017 IEEE 60th International Midwest Symposium on Circuits and Systems (MWSCAS), Medford, MA, USA, 6–9 August 2017; pp. 56–59.
- Weiss, A.C.; Kempe, K.; Förster, S.; Caruso, F. Microfluidic Examination of the “Hard” Biomolecular Corona Formed on Engineered Particles in Different Biological Milieu. Biomacromolecules 2018, 19, 2580–2594.
- Digiacomo, L.; Palchetti, S.; Giulimondi, F.; Pozzi, D.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. The biomolecular corona of gold nanoparticles in a controlled microfluidic environment. Lab Chip 2019, 19, 2557–2567.
- Palchetti, S.; Colapicchioni, V.; Digiacomo, L.; Caracciolo, G.; Pozzi, D.; Capriotti, A.L.; La Barbera, G.; Laganà, A. The protein corona of circulating PEGylated liposomes. Biochim. Biophys. Acta BBA Biomembr. 2016, 1858, 189–196.
- Palchetti, S.; Pozzi, D.; Capriotti, A.L.; La Barbera, G.; Chiozzi, R.Z.; Digiacomo, L.; Peruzzi, G.; Caracciolo, G.; Laganà, A. Influence of dynamic flow environment on nanoparticle-protein corona: From protein patterns to uptake in cancer cells. Colloids Surf. B Biointerfaces 2017, 153, 263–271.




