
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Céline Hervieu | + 3760 word(s) | 3760 | 2021-04-14 05:39:19 | | | |
| 2 | Peter Tang | Meta information modification | 3760 | 2021-06-09 11:36:25 | | |
Video Upload Options
Cancer stem cells (CSCs) fuel tumor growth, metastasis and resistance to therapy in colorectal cancer (CRC). These cells therefore represent a promising target for the treatment of CRC. This review addresses the complexity of studying CSCs in CRC research and developing clinically effective treatments to enable CRC patients to achieve a short and long-term therapeutic response.
1. Introduction
Colorectal cancer (CRC) is the fourth leading cause of cancer-related death worldwide [1]. While the occurrence and mortality rates of CRC is declining in the European countries, these rates are increasing in rapidly transitioning countries, such as many African and South Asian countries [2]. The tumor–node–metastases (TNM) classification allows the stratification of patient groups according to the stage of the disease, based on anatomical information [3][4]. The location and stage of the tumor enable both the assessment of the patient’s prognosis and the determination of the therapeutic approach, depending on the patient’s overall health as well as the status of the tumor in terms of mutation and mismatch repair (MMR) [1][5]. Therapeutic options for the treatment of CRC are surgical resection, systemic therapy including chemotherapy, targeted therapy and immunotherapy, local therapy for metastases and palliative therapy [1][6]. Importantly, surgical resection is the only curative treatment, if all macroscopic and microscopic tumor foci can be removed [1][6]. Unfortunately, even after well directed curative treatment, some patients experience treatment failure that may be associated with the development of multidrug resistance (MDR) during or after treatment. In addition, despite initially successful therapy, the development of drug resistance often leads to relapse in cancer patients, known as minimal residual disease (MRD) [7]. Both MDR and MRD can be attributed to a subpopulation of tumor cells with self-renewal and multi-lineage differentiation capabilities, the cancer stem cells (CSCs), known as colorectal cancer stem cells (CCSCs) for CRC [8]. CSCs contribute to tumor initiation and dissemination, treatment resistance and metastasis development. Tumor microenvironment (TME) and metabolic plasticity may also be involved in therapeutic failure by imposing selective pressures on cancer cells that lead to chemoresistance and cancer progression [9][10]. Therefore, the development of new therapies targeting CSCs, taking into account the TME and tumor metabolism, represents an interesting approach to overcome resistance to therapies [11].
2. Colorectal Cancer Stem Cells
The CSC theory suggests that tumor growth is driven by a small number of dedicated stem cells (SCs), the CSCs [8]. By definition, a CSC has the ability to self-renew in order to expand its pool and to generate all the differentiated cells that comprise the tumor (multi-potency). The transformation of a colorectal stem cell into CCSC requires the acquisition of tumor-related features.
2.1. Colorectal Cancer Stem Cell Origin
The history of CSCs began two decades ago with the discovery of CSCs in human acute myeloid leukemia (AML) by Dick and colleagues [12]. For the first time, a cell capable of initiating human AML in immunodeficient mice and possessing differentiation, proliferation and self-renewal capabilities was described. A few years later, using similar experimental approaches, the presence of CSC was demonstrated in solid cancers such as colorectal cancer. The origin of CSCs in CRC is controversial, and several hypotheses have been proposed. CCSCs are associated with the acquisition of malignant molecular and cellular changes either due to the accumulation of genetic and epigenetic alterations in restricted stem/progenitor cells and normal tumor cells, or to the dedifferentiation of somatic cells caused by various genetic and environmental factors [13][14][15]. CSCCs exhibit tumor-related characteristics such as uncontrolled growth, tumorigenicity and therapy resistance, and may constitute the small reservoir of drug-resistant cells that are responsible for relapses after chemotherapy-induced remission, known as MRD, and distant metastasis [7][11]. Thus, CCSCs play a key role in the initiation, invasion and progression of CRC as well as resistance to therapy. These CCSCs give rise to heterogeneous tumors that can be serially transplanted into immunodeficient mice that resemble the original tumor [16]. In addition, CCSCs have the ability to form disseminated metastatic tumors due to their extensive proliferative potential [15]. One of the main challenges in the study of CCSCs is their isolation, due to their low percentage within the tumor [16]. However, the CCSC population appears to be phenotypically and functionally heterogeneous and dynamic, which is another barrier to their isolation [17]. Therefore, the development of therapies that selectively eradicate CCSCs offers promising opportunities for a sustainable clinical response but requires effective technologies to detect and isolate them [11].
2.2. Colorectal Cancer Stem Cell Isolation Methods
Different methods are used to isolate CCSCs, based either on the expression pattern of CCSC markers, the functional aspect of CCSCs, or their biophysical features [18]. The objective of this chapter is to present the techniques currently in use with the advantages and disadvantages of each approach.
2.2.1. CCSC Isolation Based on Phenotypic Features
Many stem cells markers were found to be associated with CCSC features. However, the heterogeneous and dynamic nature of CCSCs challenges their isolation and enrichment. The first publications from the literature identifying subpopulations of CSCs in CRC are summarized in Table 1. Experimental models, CCSC isolation methods and characterization techniques used by the authors are detailed in this table. Studies conducted by O’Brien et al. and Ricci-Vitiani et al. identified the first CCSC marker: the five-transmembrane glycoprotein CD133 [19][20]. However, its use has become controversial as the tumorigenic and clonogenic potential of CD133+-CSCs depends on the positivity for a specific glycosylated epitope of the CD133 protein [21].
Table 1. Experimental models, markers and CCSC isolation and characterization methods used in the first publications identifying CSCs in CRC.
|
References |
Experimental Models |
Identified CCSC Subpopulations |
CCSC Isolation Methods |
CCSC CharacterizationAssays |
|---|---|---|---|---|
|
O’Brien et al. [20] |
CRC patient tissues CRC cells from patient tumors Animal model (mice) |
CD133+ |
MACS and FACS |
Flow cytometry Immunohistochemistry Tumorigenicity assay |
|
Ricci-Vitiani et al. [19] |
CRC patient tissues CRC cells from patient tumors Primary tumor cell cultures Animal model (mice) |
CD133+ |
MACS and FACS |
Sphere formation assay Flow cytometry Immunohistochemistry Tumorigenicity assay |
|
Dalerba et al. [22] |
CRC patient tissues CRC xenograft lines Single-cell suspensions |
EpCAMhigh/CD44+ EpCAMhigh/CD44+/CD166+ |
FACS |
ALDH assay Flow cytometry Tumorigenicity assay |
|
Barker et al. [23] |
Animal model (Ah-cre/Apcflox/flox and Lgr5-EGFP-IRES-creERT2/APCflox/flox mice) |
Lgr5+ |
/ |
LacZ analysis Immunohistochemistry |
|
Sangiorgi and Capecchi [24] |
Animal model (Bmi1-IRES-Cre-ER mice) |
Bmi1+ |
/ |
LacZ analysis Immunohistochemistry |
|
Vermeulen et al. [25] |
CRC patient tissues CRC cells and single-cell-derived cultures from patient tumors Animal model (mice) |
CD133+/CD24+ CD44+/CD166+ CD24+/CD29+ |
MACS and FACS |
Sphere formation assay In vitro differentiation assay Immunohistochemistry Flow cytometry Tumorigenicity assay |
|
Pang et al. [26] |
CRC patient tissues CRC cells from patient tumors Animal model (mice) |
CD133+/CD26+ CD133+/CD26+/CD44+ |
MACS and FACS |
Sphere formation assay In vitro invasion assays Chemotherapeutic treatments Tumorigenicity assay |
|
Todaro et al. [27] |
CRC patient tissues Sphere-derived adherent cultures CRC cells from patient tumors or spheres Animal model (mice) |
CD44v6+ |
MACS and FACS |
Immunofluorescence Immunohistochemistry Invasion assay Sphere formation assay Tumorigenicity assay |
CRC: colorectal cancer; CCSC: colorectal cancer stem cells; CD: cluster of differentiation; MACS: magnetic-activated cell sorting; FACS: fluorescence-activated cell sorting; ALDH: aldehyde dehydrogenase.
Then, Clarke’s group showed that EpCAMhigh/CD44+cells isolated from human CRC could establish a tumor in mice with morphological and phenotypic heterogeneity of the original tumor and concluded that CD44 and EPCAM markers could be considered robust CCSC markers [22]. In addition, the study by Dalerba et al. highlights an additional differentially expressed marker, CD166, which could be used to further enrich CCSCs in the EpCAhigh/CD44+ population [22]. Using lineage-tracing experiments in mice, Clevers and coworkers identified stem cells in the small intestine and colon using the marker gene Lgr5 [28] and proposed them as the cells-of-origin of intestinal cancer [23]. At the same time, Sangiorgi and Capecchi’s study found another intestinal stem cell marker in vivo, Bmi1 [24]. Importantly, Bmi-1 and Lgr5 markers define two types of SCs, quiescent and rapidly cycling SCs, respectively [23][24], and may identify CCSCs. Vermeulen et al. showed that spheroid cultures from primary CRC have a tumor-initiating capacity and that a cell subpopulation expresses CD24, CD29, CD44 and CD166 markers, suggested as CCSC markers [25]. The study by Pang et al. identifies a subpopulation of CD26+ cells capable of developing distant metastases when injected into the mouse cecal wall and associated with increased invasiveness and chemoresistance, whereas CD26− cells cannot [26]. Interestingly, the presence of CD26+ cells in the primary tumor of patients without distant metastases at that time may predict future distant metastases, highlighting a critical role of CSCs in the progression of metastatic cancer and important clinical implications [26]. The transmembrane glycoprotein CD44 has several splicing variants, including CD44v6, which appears to negatively impact the prognosis of CRC patients [29][30]. Todaro et al. demonstrated that all identified CCSCs express the CD44v6 marker, which supports their migration and promotes metastasis [27]. Each of these markers has its own function and role in the prognosis of CRC, as shown in Table 2.
Table 2. Functions and roles in CRC prognosis of CCSC markers.
|
CCSC Markers |
Functions |
Roles in Prognosis of CRC |
References |
|---|---|---|---|
|
Bmi-1 |
Polycomb-repressor protein Involved in self-renewal |
High expression of Bmi-1 is associated with poor survival |
|
|
CD24 (Heat stable antigen 24) |
Cell adhesion molecule Alternative ligand of P-selectin |
Strong cytoplasmic expression of CD24 is correlated with shortened patient survival |
|
|
CD26 |
Cell adhesion glycoprotein Promote invasion and metastases |
Elevated-CD26 expression is associated with advanced tumor staging and worse overall survival |
|
|
CD29 (Integrin-β1) |
Transmembrane proteinInvolved in cell adhesion |
Overexpression of CD29 is correlated with poor prognosis and aggressive clinicopathological features |
|
|
CD44 |
Transmembrane glycoprotein Regulate cell interactions, adhesion and migration |
CD44 overexpression is associated with lymph node metastasis, distant metastases and poor prognosis |
|
|
CD44v6 |
Bind hepatocyte growth factor Promote migration and metastases |
High level of CD44v6 has an unfavorable impact on overall survival |
|
|
CD133 (Prominin-1) |
Cell surface glycoprotein Regulate self-renewal and tumor angiogenesis |
CD133 expression is correlated with low survival in CRC patients |
|
|
CD166 (Activated leukocyte adhesion molecule) |
Cell adhesion molecule Mediate homophilic interactions |
Overexpression of CD166 is correlated with shortened patient survival |
|
|
EpCAM (Epithelial cell adhesion molecule) |
Transmembrane glycoprotein Regulate cell adhesion, proliferation and migration |
Loss of EpCAM expression is associated with tumor stage, lymph node and distant metastases and poor prognosis |
|
|
Lgr5 (Leucine-rich repeat- containing G-protein coupled receptor 5) |
Seven-transmembrane protein Target of Wnt pathway involved in self-renewal |
Lgr5 expression is associated with lymph node and distant metastases, and overexpression with reduced overall survival |
CCSC: colorectal cancer stem cells; CD: cluster of differentiation; ECM: extracellular matrix; CRC: colorectal cancer.
All these markers can be expressed by CCSCs, but they do not all have the same capacity. Some, such as CD133, Lgr5, Bmi-1, CD26 and CD44v6 alone identify CCSCs, while the other presented markers allow the identification of CCSCs only in combination with one or more of the aforementioned markers. In conclusion, these markers play a key role in the identification of CCSCs and can be used alone or in combination to sort CCSCs by magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS) techniques.
MACS is a magnetic-based cell isolation technique, using a positive selection strategy, presented in Figure 1 panel 1 [44]. Magnetic beads are conjugated to highly specific monoclonal antibodies that recognize CCSC marker on the surface of cells of interest. Then, the heterogeneous suspension of cells is passed through a separation column, in a magnetic field, to retain the cells labeled with magnetic beads and antibodies [45]. By switching off the magnetic field, target cells will be eluted. MACS is a fast and easy method of cell separation, especially for the isolation of CCSCs that represent a small cell population in the tumor mass. However, MACS is only a mono-parameter separation method that requires cell labelling and is unable to separate cells based on the variable expression of markers [44][45].
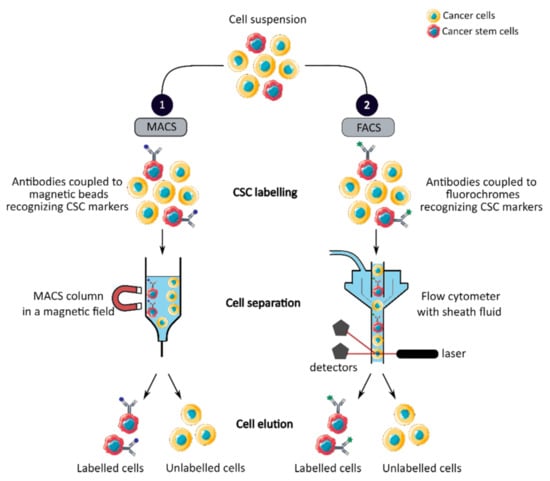
Figure 1. Phenotypic sorting of CSCs through the expression of CSC markers recognized by antibodies coupled to either magnetic beads, MACS (1), or fluorochromes, FACS (2). Once the antibodies are added, the cell suspension is passed through either a MACS column in a magnetic field that retains the antibody-labeled cells (1) or through a flow cytometer that distinguishes and isolates labeled cells from unlabeled cells (2). CSC: cancer stem cell; MACS: magnetic-activated cell sorting; FACS: fluorescence-activated cell sorting.
FACS uses fluorescently labeled antibodies that target the cell surface or intracellular markers to isolate CCSCs [44]. Antibodies are conjugated to fluorochromes and recognize the marker of interest within a cell suspension, as shown in Figure 1 panel 2 [44]. The cell suspension is then hydrodynamically focused into a stream of individual cells by the flow cytometer and passed through a laser which provides information on the size, granularity and fluorescent properties of single cells [18]. Fluorochromes with different emission wavelengths can be used simultaneously to allow multiparameter separations [44]. Both technologies allow the sorting of CCSCs with high purity but require the availability of antibodies and cell labeling, which can modify their properties and induce cell differentiation [16][44][46]. In addition, phenotypic characterization is insufficient to define a CCSC because these markers are also expressed by normal SCs.
Therefore, in order to confirm the detection and isolation of CCSCs, their functional capabilities need to be evaluated by in vitro and in vivo assays [18].
2.2.2. CCSC Isolation Based on Functional Features
CCSCs have many intrinsic properties that can be used to identify them, such as their capacity for self-renewal, multi-lineage differentiation, detoxification due to aldehyde dehydrogenase 1 (ALDH1) activity and dye exclusion ability, colony/sphere formation and tumorigenicity, which are illustrated in Figure 2. These functional characteristics have been used to develop effective methods for isolating CCSCs. The ALDH activity assay is based on the use of a fluorescent and non-toxic ALDH substrate that freely diffuses into intact and viable cells [47]. Then, in the presence of the detoxifying enzyme ALDH, the substrate is converted into a negatively charged fluorescent product that is retained inside the cells. Thus, cells with high ALDH activity become brightly fluorescent and can be measured by flow cytometry as presented in Figure 2 panel 1a [47][48]. CCSCs increase their ALDH1 activity to resist to chemotherapeutic agents and prevent apoptosis by maintaining low levels of reactive oxygen species [47]. The advantage of the ALDH assay is high stability compared to the use of surface markers, but its specificity is low due to its expression in both normal SCs and CSCs [48].
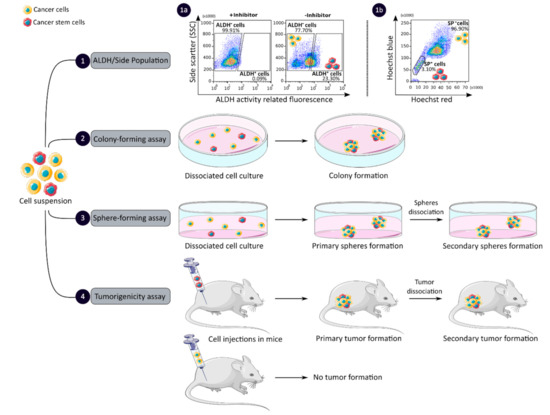
Figure 2. Functional sorting of CSCs due to their specific properties such as enhanced detoxification (1), ALDH (1a) and SP (1b), in vitro self-renewal and differentiation capacity, colony- (2) and sphere-forming (3) assays, and the ability to form tumors in vivo, tumorigenicity assay (4). CSC: cancer stem cell; ALDH: aldehyde dehydrogenase; SP: side population.
The side population (SP) assay relies on the differential ability of the cells to efflux dye via ATP-binding cassette (ABC) transporters [49]. Hoechst33342 is a fluorescent dye that binds all nucleic acids and has the particularity of passing through the plasma membrane of living cells. When excited by UV lights, Hoechst dye emits a fluorescence that can be detected by a flow cytometer [49]. SP cells are capable of actively removing the dye from the cell and have a unique low Hoechst fluorescence emission, as shown in Figure 2 panel 1b. CCSCs highly express efflux transporters, such as multidrug resistance protein 1 (ABCB1), multidrug resistance-associated proteins (ABCC1) and breast cancer resistance protein (ABCG2), to protect themselves against cytotoxic substances and therefore look like SP cells [18]. The SP assay is an easy and reliable method that does not require cell labeling, but due to its low purity and specificity, the SP assay is often combined with cell labeling to significantly increase the purity of sorted CSCCs [18][49].
Colony and sphere formation assays evaluate in vitro the self-renewal and differentiation capacities of individual cells in two (2D) and three (3D) dimensions, respectively, which are shown in Figure 2, panels 2 and 3 [50][51]. Both assays are based on non-adherent cultures using either a soft agar layer (2D) or low adherent plates (3D) [52][53]. In the soft agar method illustrated in Figure 2 panel 2, the suspension of individual cells is mixed with the soft agar which may, after several weeks of incubation, give colonies that can be stained with crystal violet to determine their number and size [50][52]. In comparison, in the 3D culture shown in Figure 2 panel 3, the individual cells in suspension are grown at very low cell density and in serum-free medium (DMEM/F12 medium) supplemented with growth factors (human recombinant basic fibroblast growth factor and human recombinant epidermal growth factor), N2 supplement, glucose, insulin and optionally antibiotics such as penicillin/streptomycin for several weeks to obtain spheroids [51][54]. The produced spheroids mimic various characteristics of solid tumors, such as growth kinetics, gene expression pattern and cellular organization with the outer layer containing highly proliferative cells, the middle layer with senescent or quiescent cells and the inner layer comprising necrotic cells due to a lack of oxygen and nutrients [53]. CCSCs can be identified in both techniques as they have the ability to form larger and more numerous colonies and are capable of giving rise to a tumor sphere (colonosphere) resembling the primary sphere when passed in series, due to their ability to grow and divide independently of their environment which normal cells are unable to do because of anoikis [18][52][55]. Thus, in vitro, 3D models appear to be a relevant preclinical model for testing new drugs, evaluating potential combinations and understanding drug resistance, by mimicking CSC-containing tumors in vitro, before testing them in vivo [18][53][55]. However, these models require well-established protocols and appropriate cell dilution to certify that each colony/sphere is derived from a single cell [18].
The tumorigenicity assay is considered the gold standard method for studying the CSC properties of human tumors in vivo [18][56]. This approach allows to determine the tumor-initiating ability of cancer cells in immunodeficient mice and their capacity for self-renewal in vivo after the dissociation of primary tumors and transplantation in secondary recipient mice, as illustrated in Figure 2 panel 4 [57]. In vivo limiting dilution is the best method for identifying the lowest concentration of cells capable of forming a tumor and determining the frequency of CSCs [18][58]. Importantly, only CSCs have the ability to generate a xenograft that is histologically similar to the parental tumor from which it originated, to be serially transplanted in a xenograft assay due to their long-term self-renewal capacity, and to generate daughter cells [56][58]. However, the use of mouse models requires ethical consideration and complicated laboratory equipment. In addition, the results of xenograft experiments are highly dependent on the number of cells, the implantation site and the incubation period, which leads to certain limitations [18]. Nevertheless, mouse models remain unique models for studying the biology of CSCs in vivo [57][58].
2.2.3. CCSC Isolation Based on Biophysical Features
The development of enrichment and isolation methods for CCSCs without cell labeling offers new perspectives, such as sorting techniques based on biophysical characteristics. The sedimentation field-flow fractionation (SdFFF) is a gentle, non-invasive and label-free method that prevents interference for further cell use and the allows separation of cells according to their size, density, shape and rigidity [16][59]. Cell separation by SdFFF depends on the differential elution of cell subpopulations submitted both to the action of a parabolic profile generated by the mobile phase in the channel and to a multigravitational external field generated by the rotation of the channel, as presented in Figure 3 [16][59]. In the past decade, SdFFF cell sorting has been adapted and applied in many fields such as neurology, oncology and stem cells [16][60][61][62]. The study by Mélin et al. describes a strategy, based on SdFFF elution, to obtain activated and quiescent CSC subpopulations from eight different human CRC cell lines [16]. The combination of cell sorting by SdFFF with the grafting of these CSC-enriched fractions into chick embryo chorioallantoic membrane (CAM) model demonstrates the potential of SdFFF to produce innovative matrices for the study of carcinogenesis and the analysis of treatment sensitivity [16][63]. The advantages of this isolation method are the use of biophysical characteristics for cell sorting without cell labeling; however, this technique requires a large number of cells and is time consuming [46].
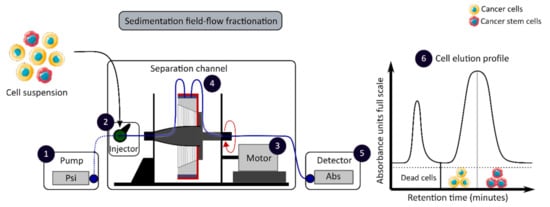
Figure 3. Biophysical sorting of CSCs according to their size, density, shape and rigidity using the SdFFF technique, which does not require cell labelling or fixation. The SdFFF is composed of a pump (1) to transport the mobile phase (PBS) and the cells, an injector (2) to introduce the cell suspension, a motor (3) to rotate the separation channel (4) and a detector (5) coupled to a computer to obtain the elution profile of the cell suspension (6). Psi is a common unit of pressure. CSC: cancer stem cell; SdFFF: sedimentation field-flow fractionation; PBS: phosphate-buffered saline; Abs: absorbance.
2.2.4. CCSC Isolation Methods: Discussion
Taken together, this chapter provides an overview of the techniques commonly used to identify and sort CCSCs, which are summarized in Table 3. The use of cell surface markers remains the most widely used in cancer research, however, it remains controversial due to the lack of a universal marker for CCSCs. Moreover, nowadays, none of the CSC isolation techniques are capable of 100% enrichment of CCSCs due to the shared properties between normal SCs, non-CCSCs and CCSCs [14][17]. As an example, Shmelkov and colleagues have shown that CD133 expression in the colon is not limited to SCs but is also expressed on differentiated tumors cells [64]. In addition, the authors found that both CD133+ and CD133− isolated from metastatic colon tumors are capable of initiating tumors in a serial xenotransplantation model [64]. A few years later, the study by Kemper et al. demonstrated that CD133 is expressed on the cell surface of CSCs and differentiated tumor cells but is differentially glycosylated [21]. Similarly, using the ALDH activity assay, Huang et al. found that ALDH1 is a marker of both normal and malignant human colonic SCs [48]. Consequently, cell surface markers and ALDH activity cannot be used alone to sort and define CSCs. Thus, the SdFFF technique offers new perspectives for CSC sorting that does not require cell labeling or fixation and thereby allows the combination of this technique with other CSC characterization methods. Therefore, the combined use of CCSC isolation methods can provide a more powerful and efficient tool for identifying and sorting CCSCs. The advantages and weaknesses of each method must be known in order to select the best method based on the experimental question, as shown in Table 3.
Table 3. Advantages and disadvantages of CCSC isolation methods.
|
Features |
Isolation Methods |
Advantages |
Disadvantages |
References |
|---|---|---|---|---|
|
Phenotypic |
MACS |
High specificity Fast and easy method |
No universal CCSC marker Monoparameter separation |
|
|
FACS |
High specificity Multiparameter separation |
No universal CCSC marker Require large number of cells |
||
|
Functional |
ALDH activity assay |
High stability |
Low specificity |
|
|
Side population assay |
No cell labelling required |
Low purity and specificity |
[49] |
|
|
Colony and sphere formation assay |
No need for complicated laboratory equipment |
Absence of standardized protocol Require proper cell dilution |
||
|
Tumorigenicity assay |
Gold standard method |
Complicated laboratory equipment Ethical consideration |
||
|
Biophysical |
SdFFF |
No cell labelling required Cell size and density separation |
Time consuming |
CCSC: colorectal cancer stem cell; MACS: magnetic-activated cell sorting; FACS: fluorescence-activated cell sorting; ALDH: aldehyde dehydrogenase; SdFFF: sedimentation field flow fractionation.
References
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal Cancer. Lancet 2019, 394, 1467–1480.
- World Health Organization: Regional Office for Europe. World Cancer Report: Cancer Research for Cancer Development; IARC: Lyon, France, 2020; ISBN 978-92-832-0447-3.
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 978-1-119-26357-9.
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: Cham, Switzerland, 2017.
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Cederquist, L.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; et al. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J. Natl. Compr. Canc. Netw. 2018, 16, 359–369.
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO Consensus Guidelines for the Management of Patients with Metastatic Colorectal Cancer. Ann. Oncol. 2016, 27, 1386–1422.
- Van der Heijden, M.; Vermeulen, L. A Cancer Stem Cell Perspective on Minimal Residual Disease in Solid Malignancies. In Cancer Stem Cell Resistance to Targeted Therapy; Maccalli, C., Todaro, M., Ferrone, S., Eds.; Resistance to Targeted Anti-Cancer Therapeutics; Springer International Publishing: Cham, Switzerland, 2019; Volume 19, pp. 31–49. ISBN 978-3-030-16623-6.
- Batlle, E.; Clevers, H. Cancer Stem Cells Revisited. Nat. Med. 2017, 23, 1124–1134.
- Desbats, M.A.; Giacomini, I.; Prayer-Galetti, T.; Montopoli, M. Metabolic Plasticity in Chemotherapy Resistance. Front. Oncol. 2020, 10, 281.
- Serpa, J. Tumor Microenvironment: The Main Driver of Metabolic Adaptation; Serpa, J., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2020; Volume 1219, ISBN 978-3-030-34024-7.
- Jordan, C.T. Cancer Stem Cells. N. Engl. J. Med. 2006, 355, 1253–1261.
- Bonnet, D.; Dick, J.E. Human Acute Myeloid Leukemia Is Organized as a Hierarchy That Originates from a Primitive Hematopoietic Cell. Nat. Med. 1997, 3, 730–737.
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Göktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal Tumorigenesis Initiated by Dedifferentiation and Acquisition of Stem-Cell-like Properties. Cell 2013, 152, 25–38.
- Jahanafrooz, Z.; Mosafer, J.; Akbari, M.; Hashemzaei, M.; Mokhtarzadeh, A.; Baradaran, B. Colon Cancer Therapy by Focusing on Colon Cancer Stem Cells and Their Tumor Microenvironment. J. Cell. Physiol. 2020, 235, 4153–4166.
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer Stem Cells (CSCs) in Cancer Progression and Therapy. J. Cell. Physiol. 2019, 234, 8381–8395.
- Mélin, C.; Perraud, A.; Akil, H.; Jauberteau, M.-O.; Cardot, P.; Mathonnet, M.; Battu, S. Cancer Stem Cell Sorting from Colorectal Cancer Cell Lines by Sedimentation Field Flow Fractionation. Anal. Chem. 2012, 84, 1549–1556.
- Hirata, A.; Hatano, Y.; Niwa, M.; Hara, A.; Tomita, H. Heterogeneity of Colon Cancer Stem Cells. In Stem Cells Heterogeneity in Cancer; Birbrair, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; Volume 1139, pp. 115–126. ISBN 978-3-030-14365-7.
- Akbarzadeh, M.; Maroufi, N.F.; Tazehkand, A.P.; Akbarzadeh, M.; Bastani, S.; Safdari, R.; Farzane, A.; Fattahi, A.; Nejabati, H.R.; Nouri, M.; et al. Current Approaches in Identification and Isolation of Cancer Stem Cells. J. Cell. Physiol. 2019, 234, 14759–14772.
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and Expansion of Human Colon-Cancer-Initiating Cells. Nature 2007, 445, 111–115.
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A Human Colon Cancer Cell Capable of Initiating Tumour Growth in Immunodeficient Mice. Nature 2007, 445, 106–110.
- Kemper, K.; Sprick, M.R.; de Bree, M.; Scopelliti, A.; Vermeulen, L.; Hoek, M.; Zeilstra, J.; Pals, S.T.; Mehmet, H.; Stassi, G.; et al. The AC133 Epitope, but Not the CD133 Protein, Is Lost upon Cancer Stem Cell Differentiation. Cancer Res. 2010, 70, 719–729.
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic Characterization of Human Colorectal Cancer Stem Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163.
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt Stem Cells as the Cells-of-Origin of Intestinal Cancer. Nature 2009, 457, 608–611.
- Sangiorgi, E.; Capecchi, M.R. Bmi1 Is Expressed in Vivo in Intestinal Stem Cells. Nat. Genet. 2008, 40, 915–920.
- Vermeulen, L.; Todaro, M.; de Sousa Mello, F.; Sprick, M.R.; Kemper, K.; Perez Alea, M.; Richel, D.J.; Stassi, G.; Medema, J.P. Single-Cell Cloning of Colon Cancer Stem Cells Reveals a Multi-Lineage Differentiation Capacity. Proc. Natl. Acad. Sci. USA 2008, 105, 13427–13432.
- Pang, R.; Law, W.L.; Chu, A.C.Y.; Poon, J.T.; Lam, C.S.C.; Chow, A.K.M.; Ng, L.; Cheung, L.W.H.; Lan, X.R.; Lan, H.Y.; et al. A Subpopulation of CD26+ Cancer Stem Cells with Metastatic Capacity in Human Colorectal Cancer. Cell Stem Cell 2010, 6, 603–615.
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356.
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of Stem Cells in Small Intestine and Colon by Marker Gene Lgr5. Nature 2007, 449, 1003–1007.
- Saito, S.; Okabe, H.; Watanabe, M.; Ishimoto, T.; Iwatsuki, M.; Baba, Y.; Tanaka, Y.; Kurashige, J.; Miyamoto, Y.; Baba, H. CD44v6 Expression Is Related to Mesenchymal Phenotype and Poor Prognosis in Patients with Colorectal Cancer. Oncol. Rep. 2013, 29, 1570–1578.
- Ma, L.; Dong, L.; Chang, P. CD44v6 Engages in Colorectal Cancer Progression. Cell Death Dis. 2019, 10, 30.
- Yanai, H.; Atsumi, N.; Tanaka, T.; Nakamura, N.; Komai, Y.; Omachi, T.; Tanaka, K.; Ishigaki, K.; Saiga, K.; Ohsugi, H.; et al. Intestinal Cancer Stem Cells Marked by Bmi1 or Lgr5 Expression Contribute to Tumor Propagation via Clonal Expansion. Sci. Rep. 2017, 7, 41838.
- Du, J.; Li, Y.; Li, J.; Zheng, J. Polycomb Group Protein Bmi1 Expression in Colon Cancers Predicts the Survival. Med. Oncol. 2010, 27, 1273–1276.
- Weichert, W. Cytoplasmic CD24 Expression in Colorectal Cancer Independently Correlates with Shortened Patient Survival. Clin. Cancer Res. 2005, 11, 6574–6581.
- Lam, C.S.-C.; Cheung, A.H.-K.; Wong, S.K.-M.; Wan, T.M.-H.; Ng, L.; Chow, A.K.-M.; Cheng, N.S.-M.; Pak, R.C.-H.; Li, H.-S.; Man, J.H.-W.; et al. Prognostic Significance of CD26 in Patients with Colorectal Cancer. PLoS ONE 2014, 9, e98582.
- Liu, Q.-Z.; Gao, X.-H.; Chang, W.-J.; Gong, H.-F.; Fu, C.-G.; Zhang, W. Expression of ITGB1 Predicts Prognosis in Colorectal Cancer: A Large Prospective Study Based on Tissue Microarray. Int. J. Clin. Exp. Pathol. 2015, 8, 12802.
- Du, L.; Wang, H.; He, L.; Zhang, J.; Ni, B.; Wang, X.; Jin, H.; Cahuzac, N.; Mehrpour, M.; Lu, Y.; et al. CD44 Is of Functional Importance for Colorectal Cancer Stem Cells. Clin. Cancer Res. 2008, 14, 6751–6760.
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell. Physiol. Biochem. 2018, 46, 860–872.
- Wang, Z.; Tang, Y.; Xie, L.; Huang, A.; Xue, C.; Gu, Z.; Wang, K.; Zong, S. The Prognostic and Clinical Value of CD44 in Colorectal Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 309.
- Zhu, L.; Gibson, P.; Currle, D.S.; Tong, Y.; Richardson, R.J.; Bayazitov, I.T.; Poppleton, H.; Zakharenko, S.; Ellison, D.W.; Gilbertson, R.J. Prominin 1 Marks Intestinal Stem Cells That Are Susceptible to Neoplastic Transformation. Nature 2009, 457, 603–607.
- Glumac, P.M.; LeBeau, A.M. The Role of CD133 in Cancer: A Concise Review. Clin. Transl. Med. 2018, 7, 18.
- Weichert, W. ALCAM/CD166 Is Overexpressed in Colorectal Carcinoma and Correlates with Shortened Patient Survival. J. Clin. Pathol. 2004, 57, 1160–1164.
- Han, S.; Zong, S.; Shi, Q.; Li, H.; Liu, S.; Yang, W.; Li, W.; Hou, F. Is Ep-CAM Expression a Diagnostic and Prognostic Biomarker for Colorectal Cancer? A Systematic Meta-Analysis. EBioMedicine 2017, 20, 61–69.
- Han, Y.; Xue, X.; Jiang, M.; Guo, X.; Li, P.; Liu, F.; Yuan, B.; Shen, Y.; Guo, X.; Zhi, Q.; et al. LGR5, a Relevant Marker of Cancer Stem Cells, Indicates a Poor Prognosis in Colorectal Cancer Patients: A Meta-Analysis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 267–273.
- Cammareri, P.; Lombardo, Y.; Francipane, M.G.; Bonventre, S.; Todaro, M.; Stassi, G. Isolation and Culture of Colon Cancer Stem Cells. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 86, pp. 311–324. ISBN 978-0-12-373876-9.
- Korkusuz, P.; Köse, S.; Yersal, N.; Önen, S. Magnetic-Based Cell Isolation Technique for the Selection of Stem Cells. In Skin Stem Cells; Turksen, K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1879, pp. 153–163. ISBN 978-1-4939-8869-3.
- Mathonnet, M. Hallmarks in Colorectal Cancer: Angiogenesis and Cancer Stem-like Cells. World J. Gastroenterol. 2014, 20, 4189.
- Mele, L.; Liccardo, D.; Tirino, V. Evaluation and Isolation of Cancer Stem Cells Using ALDH Activity Assay. In Cancer Stem Cells; Papaccio, G., Desiderio, V., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1692, pp. 43–48. ISBN 978-1-4939-7400-9.
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde Dehydrogenase 1 Is a Marker for Normal and Malignant Human Colonic Stem Cells (SC) and Tracks SC Overpopulation during Colon Tumorigenesis. Cancer Res. 2009, 69, 3382–3389.
- Golebiewska, A.; Brons, N.H.C.; Bjerkvig, R.; Niclou, S.P. Critical Appraisal of the Side Population Assay in Stem Cell and Cancer Stem Cell Research. Cell Stem Cell 2011, 8, 136–147.
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic Assay of Cells in Vitro. Nat. Protoc. 2006, 1, 2315–2319.
- Shaheen, S.; Ahmed, M.; Lorenzi, F.; Nateri, A.S. Spheroid-Formation (Colonosphere) Assay for in Vitro Assessment and Expansion of Stem Cells in Colon Cancer. Stem Cell Rev. Rep. 2016, 12, 492–499.
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The Soft Agar Colony Formation Assay. J. Vis. Exp. 2014, 51998.
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441.
- Relier, S.; Yazdani, L.; Ayad, O.; Choquet, A.; Bourgaux, J.-F.; Prudhomme, M.; Pannequin, J.; Macari, F.; David, A. Antibiotics Inhibit Sphere-Forming Ability in Suspension Culture. Cancer Cell Int. 2016, 16, 6.
- Visvader, J.E.; Lindeman, G.J. Cancer Stem Cells in Solid Tumours: Accumulating Evidence and Unresolved Questions. Nat. Rev. Cancer 2008, 8, 755–768.
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer Stem Cells: An Evolving Concept. Nat. Rev. Cancer 2012, 12, 133–143.
- Aiken, C.; Werbowetski-Ogilvie, T. Animal Models of Cancer Stem Cells: What Are They Really Telling Us? Curr. Pathobiol. Rep. 2013, 1, 91–99.
- O’Brien, C.A.; Kreso, A.; Jamieson, C.H.M. Cancer Stem Cells and Self-Renewal. Clin. Cancer Res. 2010, 16, 3113–3120.
- Mélin, C.; Perraud, A.; Bounaix Morand du Puch, C.; Loum, E.; Giraud, S.; Cardot, P.; Jauberteau, M.-O.; Lautrette, C.; Battu, S.; Mathonnet, M. Sedimentation Field Flow Fractionation Monitoring of in Vitro Enrichment in Cancer Stem Cells by Specific Serum-Free Culture Medium. J. Chromatogr. B 2014, 963, 40–46.
- Lacroix, A.; Deluche, E.; Zhang, L.Y.; Dalmay, C.; Mélin, C.; Leroy, J.; Babay, M.; Morand Du Puch, C.; Giraud, S.; Bessette, B.; et al. A New Label-Free Approach to Glioblastoma Cancer Stem Cell Sorting and Detection. Anal. Chem. 2019, 91, 8948–8957.
- Vedrenne, N.; Sarrazy, V.; Battu, S.; Bordeau, N.; Richard, L.; Billet, F.; Coronas, V.; Desmoulière, A. Neural Stem Cell Properties of an Astrocyte Subpopulation Sorted by Sedimentation Field-Flow Fractionation. Rejuvenation Res. 2016, 19, 362–372.
- Faye, P.-A.; Vedrenne, N.; De la Cruz-Morcillo, M.A.; Barrot, C.-C.; Richard, L.; Bourthoumieu, S.; Sturtz, F.; Funalot, B.; Lia, A.-S.; Battu, S. New Method for Sorting Endothelial and Neural Progenitors from Human Induced Pluripotent Stem Cells by Sedimentation Field Flow Fractionation. Anal. Chem. 2016, 88, 6696–6702.
- Mélin, C.; Perraud, A.; Christou, N.; Bibes, R.; Cardot, P.; Jauberteau, M.-O.; Battu, S.; Mathonnet, M. New Ex-Ovo Colorectal-Cancer Models from Different SdFFF-Sorted Tumor-Initiating Cells. Anal. Bioanal. Chem. 2015, 407, 8433–8443.
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; St. Clair, R.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 Expression Is Not Restricted to Stem Cells, and Both CD133+ and CD133− Metastatic Colon Cancer Cells Initiate Tumors. J. Clin. Investig. 2008, JCI34401.




