Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Milva Pepi | + 4282 word(s) | 4282 | 2021-06-01 06:22:08 |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pepi, M.; Focardi, S. Antibiotic-Resistant Bacteria and Aquaculture. Encyclopedia. Available online: https://encyclopedia.pub/entry/10618 (accessed on 07 February 2026).
Pepi M, Focardi S. Antibiotic-Resistant Bacteria and Aquaculture. Encyclopedia. Available at: https://encyclopedia.pub/entry/10618. Accessed February 07, 2026.
Pepi, Milva, Silvano Focardi. "Antibiotic-Resistant Bacteria and Aquaculture" Encyclopedia, https://encyclopedia.pub/entry/10618 (accessed February 07, 2026).
Pepi, M., & Focardi, S. (2021, June 08). Antibiotic-Resistant Bacteria and Aquaculture. In Encyclopedia. https://encyclopedia.pub/entry/10618
Pepi, Milva and Silvano Focardi. "Antibiotic-Resistant Bacteria and Aquaculture." Encyclopedia. Web. 08 June, 2021.
Copy Citation
Antibiotic residues originated from aquaculture can select for resistant aquatic bacteria, promoting the spread of antibiotic resistance, even when concentrations were below the minimum inhibitory concentration (MIC) of bacterial strains of the community.
aquaculture
antibiotic-resistance
Mediterranean Sea
1. Induction of Antibiotic Resistance
Antibiotic residues can select for resistant aquatic bacteria, promoting the spread of antibiotic resistance, even when concentrations were below the minimum inhibitory concentration (MIC) of bacterial strains of the community [1][2][3]. High frequencies of antibiotic-resistant bacteria have been reported in sites near aquaculture where antibiotics have been used, demonstrating that modified antibiotics in an aquaculture facility have a high potential to exert selective pressure and increase the frequency of antibiotic resistance in other environmental bacteria [4][5]. In the aquatic environment, 90% of aquatic bacteria show resistance to at least one antibiotic, and approximately 20% were multi antibiotic-resistant. In the case of simultaneous application of different antibiotics in aquaculture, multiresistant bacteria can develop. Bacteria carrying genes coding for novel antibiotic resistance mechanisms were moreover present [6]. Antibiotic resistance allows bacteria the survive high concentrations of antibiotics, conferring a selective advantage to members of communities carrying resistances. Resistant bacterial strains prevail over susceptible ones. An important and at the same time worrying aspect is that the antibiotics used in aquaculture include those used in human therapies, thus inducing resistance to these antibiotics [7].
In Mediterranean aquaculture facilities, misuse of antibiotics caused an increase in levels of antibiotics in the surrounding sediments and water column [8]. The use of antibiotics in the Mediterranean farmed European seabass (D. labrax) and gilthead seabream (S. aurata), describing oxytetracycline, oxolinic acid, flumequine, and potentiated sulphonamides evidencing methaphylaxis, a control treatment of a group of animals after the diagnosis of infection and/or clinical disease in part of the group, as the best practice for their use [9].
2. Transfer of Antibiotic Resistance between Bacteria
Aquaculture sites represent ‘hotspots for antibiotic-resistant genes’ [10][11][12][13]. Bacteria harboring different genes of antibiotic resistance can grow according to environmental features, spreading genes in different sites [14]. The aquatic environment may also contain human and animal bacterial pathogens, which act as agents in sharing genetic determinants between aquatic and terrestrial bacteria [15]. The set of mobile genetic elements in a genome is called a mobilome and can spread among aquatic bacteria. The mobilome comprehends naked DNA, insertion sequences, insertion sequence elements with common regions, integrons mobilized by plasmids, transposons, and integrative and conjugative elements, genomic islands, transposons and conjugative transposons, conjugative and mobilizable plasmids and bacteriophages, including phage-like elements designated gene transfer agents [15].
Horizontal gene transfer between aquatic and human pathogens is an important phenomenon involving antibiotic resistance genes. New genetic elements can thus enter into terrestrial bacterial communities, including human pathogens, the latter becoming more difficult to treat [16]. The same aquatic environments with aquaculture facilities can present unique conditions allowing horizontal gene transfer. One case is represented by the biofilms of aquatic bacteria attached to organic particulate matter, sediment clays and sands, and fish farm structures, combined with the large concentrations of bacteriophages and gene transfer agents in seawater, also allowing the transfer of horizontal gene and dissemination of antibiotic resistance [17]. In aquatic environments, horizontal gene transfer can be mediated by DNA generated by lysis of bacteriophage and by plasmids. Both naked DNA from bacteriophages and plasmids can contain antimicrobial resistance genes that may be expressed after entering bacteria. Aquatic bacteria such as Vibrio spp. resulted naturally competent for DNA uptake, allowing transformation to occur in the aquatic environment [15]. Antibiotic-resistant genes are characterized by a different persistence in the environment, depending on the plasmid or chromosomal origin. Higher mobility and higher concentrations characterize antibiotic-resistant genes originated from plasmid with respect to those from chromosomes [18].
Genes of antibiotic resistance are present as both intracellular and extracellular fractions of DNA in the environment. Intracellular and extracellular antibiotic resistance genes observe the same pathway in sediments in an aquaculture site and nearby sites, revealing the presence of connections between these different sites [19][20]. Characteristics of DNA molecules also impact the persistence of antibiotic-resistant genes in the extracellular environment [18]. Some extracellular antibiotic-resistant genes are more recalcitrant to DNA degrading enzymes (DNases) than others, likely due to their sequence and structural features [21]. Due to the higher surface charges and higher molecular flexibility, chromosomal DNA is more adsorptive than plasmid DNA [22]. Therefore, in the environment, extracellular chromosomal DNA can persist longer than extracellular plasmid DNA. In sediments, a lower detection frequency of extracellular DNA located on the plasmid is expected with respect to chromosomal DNA [23].
2.1. Conjugation
Intracellular antibiotic-resistant genes may be disseminated via conjugation from cell-to-cell contact, and transduction because of infection of bacterial phages [24]. Conjugation is a horizontal gene transfer mechanism via cell to cell contact through junction due to pili or adhesins, by using a pore [25]. Conjugation is associated with plasmids that can transfer faster than a whole chromosome [26][27]. A percentage higher than 50% of known plasmids can be transferred by using conjugation as a horizontal gene transfer mechanism [28]. The process of conjugation may be present between the same bacterial species, but may also occur between unrelated populations characterized by a taxonomic distance, while at lower frequencies [25][26]. Plasmids and transposons can facilitate the conjugative transfer of antibiotic resistance by collecting antibiotic-resistant genes and carrying them to recipient cells [25][29]. Conjugation of antibiotic-resistant genes has been frequently reported in various environments, including soil, sediments, water, food, plant, animal, and clinical bacteria [26][28][30]. Conjugative transfer of multi-drug resistance among and across bacteria in different environments was showed [23][26] (Figure 1A).
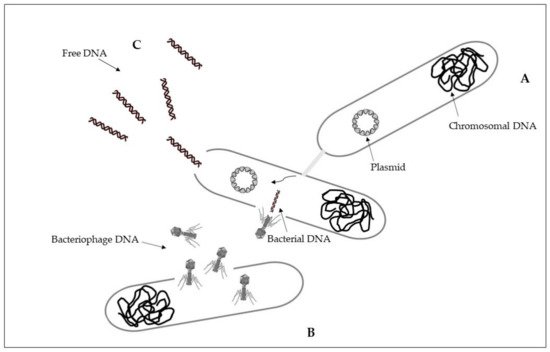
Figure 1. Horizontal gene transfer: (A), conjugation; (B), transduction; (C), transformation. See text for explanations.
2.2. Transduction
Transduction is a horizontal gene transfer mechanism with bacteriophages that act as mediators for intracellular DNA transfer from an infected bacterial cell to a recipient bacterial cell [26][31]. Bacteriophages, or phages, are viruses that infect bacteria and are able to collect and transfer genes to a host bacterial [26][32]. Bacteriophages can transfer both chromosomal DNA and plasmid DNA [31][33]. Once DNA is transferred, it must be incorporated into the recipient chromosome by homologous recombination [31]. Some bacteriophages have a wide range of bacterial hosts and can move across different species [34]. The transfer of antibiotic resistance genes via phages for different bacterial species has been extensively described [35][36][37]. During the first phase, bacteriophages attach to the bacterial host and then inject their genome that has the capability to sequester the molecular machinery of the host bacterial cell to synthesize new phages. New phages can then lyse the host cell and spread to the environment [38] (Figure 1B).
2.3. Transformation
Differently from intracellular DNA, extracellular antibiotic-resistant genes can enter the competent cells of non-resistant bacteria through the mechanism of natural transformation [24]. Extracellular DNA can originate from the lysis of dead cells and the secretion from live cells, representing a dynamic gene pool for natural transformation. Adsorption on sediment colloids, sand particles, clay minerals, and humic substances can protect extracellular DNA against enzyme nuclease attacks [39][40]. Natural transformation represents a direct uptake and integration of extracellular DNA [41]. It is essential that bacterial cells must first develop a regulated physiological state, a defined state of competence, for carrying out natural transformation [25][42]. Specific environmental conditions can stimulate competence development [26][43]. On the basis of the absorption of extracellular DNA by competent cells, there may be reasons such as nutrition, chromosomal DNA repair, and diversification of the genetic material for evolution [42]. Natural transformation requests the persistence of DNA in the extracellular environment, and the ability to resist degradation in environmental conditions. Higher efficiency of natural transformation is evident in the presence of longer extracellular DNA fragments with respect to the results obtained with smaller extracellular DNA fragments [44][45]. Once in the recipient cell, the new DNA must be integrated into the recipient bacterial genome in the case of chromosomal DNA, and be integrated or recircularized into a self-replicating plasmid, in the case of plasmid DNA [42]. Natural transformation through chromosomal DNA is thus more efficient than in the presence of plasmid DNA [23][25] (Figure 1C).
3. Transfer of Antibiotic Resistance from Aquaculture Bacteria to Human Pathogens
3.1. Quinolones
Antibiotics of the quinolones class are widely used in aquaculture, with traces found in aquaculture effluents, the water column, and sediments near aquaculture facilities. The quinolone concentrations found in these compartments are high enough to exert selective pressure on aquatic bacterial species. The latter can mix with species of different origins, favoring gene exchange and spreading resistance to antibiotics [46]. Quinolones are antibiotics with a broad spectrum against both Gram-positive and Gram-negative bacteria, including mycobacteria and anaerobes. They exert their actions by inhibiting bacterial nucleic acid synthesis through disrupting the enzymes topoisomerase IV and DNA gyrase, and by causing breakage of bacterial chromosomes. Mechanisms of resistance to quinolones provide mutations in the bacteria genes, as the mutation in genes encoding the DNA gyrase and topoisomerase IV targets, or other genomic alterations which alter topoisomerase targets, modify quinolone, or reduce drug accumulation by decreasing drug absorption or increasing drug outflow. Resistance to quinolones may result from the uptake of the plasmid gene from the environment or from other resistant strains [47]. Genes of quinolone resistance included in plasmid-mediated quinolone resistance (PMQR) are the followings: six qnr genes (qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC) encoding gyrase-protection repetitive peptides; oqxAB, qepA, and qaqBIII encoding efflux pumps; and aac(6′)-Ib-cr encoding an aminoglycoside and quinolone inactivating acetyl-transferase. Moreover, these genes can synergize with chromosomal gyrA and parA mutations conferring quinolone resistance [48]. The water-borne bacterial species Shewanella algae and Vibrio splendidus comprehend bacterial strains carrying qnrA and qnrS genes, respectively, and gene qnrS were detected in another water-borne strain, Aeromonas sp., the role of aquatic environments in the diffusion of such resistance determinants has acquired more and more importance [49]. Strains of both the aquatic bacterial species Aeromonas punctata subsp. punctata and A. media evidenced the presence of the qnrS2 gene. The qnrS2 gene was located on IncU-type plasmids in both isolates. When these plasmids were transferred into bacteria of the species Escherichia coli, they became highly resistant to quinolones and fluoroquinolones. The identification of plasmid-mediated qnr genes outside Enterobacteriaceae evidence a possible diffusion of those resistance determinants within Gram-negative bacteria [49]. The genes qnrA, qnrB, and qnrS for resistance to quinolones were found in the chromosome of marine bacteria isolated from an aquaculture facility in the Región de Los Lagos, Chile, and the same genes were detected in human pathogenic bacteria. The qnrA gene was, in fact, also found in the chromosome of two uropathogenic clinical strains of E. coli resistant to quinolones isolated from patients in a coastal area, bordering the same aquaculture region. The qnrB and qnrS genes were located in plasmids in two other E. coli strains isolated from the same clinical context [50]. Further investigations by sequencing qnrA1, qnrB1, and qnrS1 genes in quinolone-resistant E. coli and in marine bacteria, both from Chile, were identical. A horizontal gene transfer between antibiotic-resistant marine bacteria and human pathogens was confirmed [23]. Concerning genetic elements of marine bacteria and uropathogenic E. coli, both evidenced class 1 integron with similar co-linear structures, identical gene cassettes, and similarities in their flanking regions. Investigations in a Marinobacter sp. marine isolate and in an E. coli clinical isolated strain, highlight sequences immediately upstream of the qnrS gene evidencing homology to comparable sequences of numerous plasmid-located qnrS genes. These investigations confirm that horizontal gene transfer between bacteria in diverse ecological locations is facilitated [50]. PMQR can be transferred horizontally among distantly related lineages and might play a role in maintaining resistance levels in bacterial populations in the presence of sub-inhibitory concentrations of antibiotics [51][52]. The same plasmid has been shown to play an important role for the spread of resistance genes not only quinolones but also for other antibiotics such as β-lactams and aminoglycosides. In fact, qnr genes are frequently carried along with β-lactamase determinants on the same plasmids. Moreover, it was evidenced that the prevalent qnrA, qnrB, qnrS, and aac(6′)-Ib-cr genes among quinolone and cephalosporin-resistant clinical isolates of Klebsiella pneumoniae, are in the association between PMQR genes with resistance to quinolones, cephalosporins, and aminoglycosides [53]. Aquaculture is a possible source of aac(6′)-Ib-cr and qnrB2 in aquatic environments and Enterobacteriaceae were important hosts of these two genes. The ubiquitous bacteria, Aeromonas spp., served as vectors for qnrS2 by means of IncQ-type plasmids. A water-human transmission by and via Aeromonas species was evidenced [54], and a qnrS-containing plasmid was identified in an Aeromonas sp. clinical isolate [55]. Before, genes qnr have only been reported in Enterobacteriaceae [56][57], with the one exception of a qnrS-containing plasmid found in environmental A. punctata subsp. punctata and A. media isolates [49]. A plasmid containing the qnrS was detected in an Aeromonas sp. clinically isolated strain for the first time [55], evidencing that a qnrS2 gene was identified in a clinical isolate that was not within the Enterobacteriaceae family. The versatility of these determinants to spread among the different bacterial species with the consequent potential risk for human health became strongly evident [55]. From this evidence, the need to control the antibiotic resistance supervision of both clinical and environmental Aeromonas isolates has emerged [58][59]. Plasmid-encoded quinolone resistance genes (qnrA, qnrB, qnrS, and aac [6′]-1b-cr) were found in E. coli and in Klebsiella [60]. Genes for quinolone resistance were detected in the aquatic genera Vibrio, Shewanella, and Aeromonas and then those genes were detected in human and animal pathogens [15]. Bacterial strains belonging to the water-borne bacterial species S. algae and V. splendidus, evidenced the presence of qnrA and qnrS genes, respectively. The gene qnrS was identified in another water-borne strain Aeromonas sp., increasing evidence of the role of aquatic environments in spreading those resistance determinants [49]. Gram-negative bacterial species of the aquatic environment may be the reservoir of plasmid-mediated Qnr-like determinants, that seem closely related to the species V. splendidus [61]. The World Health Organization (WHO) designated E. coli that are resistant to fluoroquinolones as one of the nine pathogens of international concern [62]. A description of strains of E. coli in countries in the Mediterranean area is reported in Table 1. Values of percentages of E. coli resistance to fluoroquinolones originated mostly from hospitals, nevertheless, their origin with aquatic bacteria could be a real problem for both human and animal health, and a concern for the environment.
Table 1. Escherichia coli isolated strains resistant to fluoroquinolones in countries of the Mediterranean area.
| Country | Mean Percentage (%) | Range Percentage (%) | Year | Isolated Strains | Source |
|---|---|---|---|---|---|
| Bosnia and Herzegovina | 29 | 23–35 | 2016 | 215 | CAESAR |
| Croatia | 29 | 26–32 | 2017 | 1150 | EARS-Net |
| France | 17 | 16–18 | 2017 | 13,328 | EARS-Net |
| Greece | 34 | 32–36 | 2017 | 1464 | EARS-Net |
| Italy | 47 | 46–48 | 2017 | 6945 | EARS-Net |
| Lebanon | 45 | 33–57 | 2016 | 65 | GLASS |
| Portugal | 30 | 29–31 | 2017 | 6424 | EARS-Net |
| Spain | 33 | 32–34 | 2017 | 5557 | EARS-Net |
| Tunisia | 19 | 13–29 | 2017 | 78 | GLASS |
| Turkey | 55 | 53–57 | 2016 | 3670 | CAESAR |
3.2. Tetracyclines
Tetracyclines are antibiotics inhibiting bacterial protein synthesis by preventing the association of aminoacyl tRNA with the bacterial ribosome [63]. Bacteria could use three strategies to become resistant to tetracycline: limiting the access of tetracycline to the ribosomes, altering the ribosome to prevent effective binding of tetracycline, and producing tetracycline-inactivating enzymes [64]. Tetracyclines are commonly used in human and veterinary treatment, with oxytetracycline that is permitted to be mixed with feed for fish [65]. In Japan, bacterial strains from aquaculture fish and bacteria from a close clinical facility exhibited a high similarity for genes of tetracycline resistance, suggesting that they may have originated from the same source. Laboratory experiments in which tetracycline resistance from marine strains of genera Photobacterium, Vibrio, Alteromonas, and Pseudomonas were transferred to E. coli by conjugation, confirmed the possibility of a transfer of resistance determinants from marine bacteria to bacteria inhabiting the human gut. Moreover, the same resistance gene profile in aquatic bacteria and in human clinical isolates was evidenced. The antibiotic-resistant genes for tetracycline resistance identified in fish pathogenic bacteria are common to those identified in human pathogens, and that bacteria from different environments, such as aquatic and clinical, can share the same antibiotic-resistant genes [66][65][67]. Tetracycline resistance determinants revealed in Salmonella spp. strains were detected in fish pathogens of the species Vibrio anguillarum [68][69][70]. Moreover, the DNA sequence of these antibiotic resistance determinants has an important DNA sequence similarity to a plasmid of Pasteurella piscicida, which is also a fish pathogen [69][70]. The independently evolved tetracycline-resistance determinant tetG was first discovered in aquatic bacteria [69]. In the genome of the animal pathogen Chlamydia suis, the tetC gene probably may have originated from the genome of the aquatic bacterium Aeromonas salmonicida, a pathogen of salmon [71]. The independently evolved tetracycline-resistance determinant tetG was first discovered in aquatic bacteria [69]. Tetracycline resistance genes were identified in marine sediments from resistance genes in bacterial plasmids from marine sediments that shared high identity with transposons or plasmids from human pathogens, indicating that the sediment bacteria can spread resistance genes from pathogens [72][51]. The same resistance gene profile has been described in both fish bacteria and human clinical isolates. About half of the antibiotic resistance genes identified in fish pathogens are common to those identified in human pathogens [65][67].
3.3. β-Lactams, Macrolides, Fosfomycin, Chloramphenicol, Colistin, Florfenicol
Commercial fish and seafood may act as a reservoir for multiresistant bacteria, facilitating the dissemination of antibiotic resistance genes. Broad-spectrum β-lactamase resistance genes, including blaTEM-52, blaSHV-12, as well as cmlA, tetA, aadA, sul1, sul2, and sul3 were recovered in the faecal matter from S. aurata (Gilthead seabream) [73]. The β-lactam antibiotics inhibit the last step in peptidoglycan synthesis by acylating the transpeptidase involved in cross-linking peptides to form peptidoglycan. The targets for the actions of β-lactam antibiotics are known as penicillin-binding proteins. This binding, in turn, interrupts the terminal transpeptidation process and induces loss of viability and lysis, also through autolytic processes within the bacterial cell [74]. Although bacterial resistance to β-lactams mostly expresses through the production of β-lactamases, other mechanisms are involved. Following are the mechanisms of resistance: (i) inactivation by the production of β-lactamases; (ii) decreased penetration to the target site as resistance in Pseudomonas aeruginosa; (iii) alteration of target site penicillin-binding proteins as penicillin resistance in pneumococci; (iv) efflux from the periplasmic space through specific pumping mechanisms [75]. Other examples of resistances with aquatic origin include the gene of fosfomycin resistance isolated from the aquatic environment, the widely disseminated emerging floR gene of human pathogens, and the chloramphenicol resistance genes catII, catB9 and catB2, originating respectively from aquatic bacteria of the genera Photobacterium, Vibrio, and Shewanella [15]. Additionally, the plasmid-associated colistin resistance mediated by the mcr-1 gene appears to be another transmissible antibiotic resistance determinant that might have originated in the aquaculture facilities [72][76][77]. The macrolide resistance genes mef(C) and mef(G) in Vibrio spp. and Photobacterium spp. strains appear to have an aquatic origin [78]. Indeed, resistance genes have been found on transferable plasmids and integrons in pathogenic bacterial species of the genera Aeromonas, Yersinia, Photobacterium, Edwardsiella, and Vibrio. Class 1 integrons and IncA/C plasmids have been widely identified in important fish pathogens (Aeromonas spp., Yersinia spp., Photobacterium spp., Edwardsiella spp., and Vibrio spp.) and are thought to play a major role in the transmission of antimicrobial resistance determinants in the aquatic environment. The identification of plasmids in terrestrial pathogens (Salmonella enterica serotypes, E. coli, and others) which have considerable homology to plasmid backbone DNA from aquatic pathogens suggests that the plasmid profiles of fish pathogens are extremely plastic and mobile and constitute a considerable reservoir for antimicrobial resistance genes for pathogens in diverse environments [79]. The florfenicol determinant, floR, was detected for the first time in the fish pathogen Vibrio damsela [80]. This molecular evidence strongly suggests that there was a horizontal transmission of antibiotic resistance determinants from bacteria in the aquaculture environment to a human and terrestrial veterinary pathogen [69][70]. The epidemiology of the dissemination of S. typhimurium DT104 also suggests this pathogen could have been spread by fish meal as has happened with the Salmonella Agona that originated in Peru several years ago [69][81]. This process illustrates the potential role of the transport of antibiotic-resistant bacteria as an alternative mechanism responsible for the spread of antibiotic resistance determinants from the aquatic environment to the terrestrial environment [70][82]. As another example, V. cholerae of the Latin American epidemic of cholera that started in 1992, appeared to have acquired antibiotic resistance as a result of coming into contact with antibiotic-resistant bacteria selected through the heavy use of antibiotics in the Ecuadorian shrimp industry [69][83]. Multiresistant bacteria were evidenced in aquaculture contexts, including potential human pathogens strains of the genera Vibrio, Pseudomonas, and Salmonella, evidencing the possibility to transfer genetic determinants of resistance to pathogenic bacteria, both in water and in sediments [84]. Pathogens bacterial strains in aquaculture, belonging to the genera Aeromonas, Edwardsiella, Flavobacterium, Lactococcus, Photobacterium, Pseudomonas, Renibacterium, Streptococcus, Vibrio, Yersinia, were pointed out [85].
4. Antibiotic Resistance in the Mediterranean Basin
In the Mediterranean area, fish farms of European seabass (D. labrax) and Mediterranean gilthead seabream (S. aurata) are mostly present, and antibiotics were added by feed amendments [9]. In Western Mediterranean coastal sediments from the vicinity of Ligurian Sea coastal fish facilities, Gram-negative bacterial strains have been found showing high resistance to the antibiotics ampicillin and streptomycin. Multiple resistances were found in the same strains. Antibiotic resistance patterns close to fish farming showed high incidences of quinolone, tetracycline, and penicillin-resistant bacterial populations [86]. In the Mediterranean area, antibiotic-resistant bacterial pathogens isolated from sediments near a fish farm in Greece, were compared with bacterial isolates in Italy and France, including Vibrio spp., Pseudomonas spp., Aeromonas spp., and Pasteurella piscicida. Greek, Italian and French aquaculture sites allowed isolation of bacterial strains evidencing similar antibiotic sensitivity pathways, with resistance to erythromycin, kanamycin, and streptomycin, and sensitivity to most of the other antibiotics tested. Resistance was also demonstrated for some isolates towards the potentiated sulphonamides [87]. Bacterial strains Vibrio spp., Pseudomonas spp., and Photobacterium damselae ssp. piscicida isolated from gilthead seabream (S. aurata) from an aquaculture facility in southwestern Spain, in the Mediterranean region, evidenced high levels of antibiotic resistance [88]. Bacterial strains of V. anguillarum isolated from Greek fish farms evidenced multiple antibiotic resistance [89]. Multiple antibiotic resistance pathways have been highlighted in native marine bacterial strains isolated from fish farms located along the Adriatic Sea (Italy) and identified as Vibrio spp. Resistance to tetracycline (17%), trimethoprim-sulfadiazine (7%), and trimethoprim (2%) resulted in the most frequently obtained patterns [90][91]. Pathogenic halophilic strains Vibrio parahaemolyticus and V. vulnificus evidencing resistance to lincomycin, were moreover isolated from seafood in Italy [92]. High frequencies of Aeromonas spp. contamination in S. aurata from the Italian coast was pointed out, with elevated biodiversity among isolated bacterial strains. The bacterial strains showed high resistance to sulfadiazine, amoxicillin, ampicillin, erythromycin, cephalotin, streptomycin, and trimethoprim antibiotics. It was evidenced that almost all Aeromonas spp. strains showed multiple antibiotic resistance and potentially pathogenic species for humans were included, evidencing the capability to transfer via the food chain the genes responsible for antibiotic resistance to human pathogens [93]. More than one hundred bacterial strains were isolated from samples of S. aurata in an aquaculture site in Portugal. Included in the isolates, Enterobacter spp. and Pseudomonas spp. strains resulted in resistance to ertapenem and meropenem, antibiotics used in serious clinical infections. Several antibiotics for which resistance was found in these isolates appear in the World Health Organization list of “critically important antimicrobials” and “highly important antimicrobials” for human medicine [94]. In Mediterranean aquaculture, and in particular the Greek fish farm facilities, several registered antibiotics are currently available against bacterial infections, including tetracyclines, quinolones, and fluoroquinolones, potential sulfa, penicillin, and chloramphenicol derivatives. Oxytetracycline and quinolone drugs, with oxolinic acid and flumequine, are the most widely used in Mediterranean aquaculture [8]. A comparison was made of the distribution of tetracycline resistance genes in neighboring Greek fish farms and coastal environments. The tetA and tetK genes were detected in both habitats, while the tetC and tetE genes were present in fish farms and in wastewater, and tetM was found in fish farms and coastal sites. Some isolates were obtained, highlighting the presence of resistance genes teth, tetC, tetK, and tetM in the main part. The isolates were assigned to the genera Stenotrophomonas, Acinetobacter, Pseudomonas, Bacillus, and Staphylococcus. Isolated strains showed the presence of IncP plasmids, harboring tetracycline resistance genes (i.e., teth, tetC, tetE, and tetK), and the dissemination of IncP plasmids was evidenced. Based on these results, it has been shown that tetracyclines resistance genes from seawater sites have spread in bacterial communities, via broad-spectrum host plasmids [95]. The onset of antibiotic resistance in the environment and in non-clinical environments, highlight the importance of the investigation of antibiotic resistance in environmental contexts [96]. The use of antibiotics in aquaculture can lead to the emergence of antibiotic resistance in bacteria that are pathogenic to humans, posing a serious threat to public health [97]. In 2015, antibiotic-resistant bacteria increased in the European area, causing over 33,000 human deaths. Furthermore, this number is projected to increase by 2030, due to rapid socioeconomic growth and population expansion [98][99]. The emergence of antibiotic resistance in bacteria poses a serious threat, with a reduction in the use of antibiotics in aquaculture feed that is absolutely required by 2030 [100]. In Table 3, a description of antibiotic-resistant bacteria, and eventual genes for antibiotic resistance, in aquaculture sites of different countries of the Mediterranean basin was reported. The presence of antibiotic-resistant bacteria from aquaculture facilities in the Mediterranean area, highlighting serious concerns for human and animal health and for the environment, is evidenced.
Table 2. Bacteria isolated from aquaculture in the Mediterranean area, antibiotic resistance and antibiotic-resistance genes.
| Country/Area | Isolated Bacteria Species/Genus/Family/Order/Class | Antibiotic-Resistance | Antibiotic-Resistant Genes | References |
|---|---|---|---|---|
| Algeria | Vibrio alginolyticus, V. cholerae, V. fluvialis, V. hollisae | [133] | ||
| Croatia | Aeromonas spp. | [134] | ||
| Eastern Adriatic | Vibrio spp. | flumequinone, chloramphenicol, oxytetracycline | [135] | |
| Egypt | Aeromonas spp. | chloramphenicol, kanamycin, azithromycin | [136] | |
| Egypt | Pseudomonas anguilliseptica | [137] | ||
| Egypt | Aeromonas hydrophila | penicillin, erythromycin | [138] | |
| Egypt | Enterobacteriaceae | cephalosporins, carbapenem | blaKPC (blaCTX-M-15, blaSHV, blaTEM, blaPER-1) | [139] |
| France | Flavobacterium psychrophilum | [140] | ||
| France | Yersinia ruckeri | [141] | ||
| Greece | Acinetobacter spp., Bacillus spp., Pseudomonas spp., Staphylococcus spp., Stenotrophomonas spp. | tetracycline | tetA, tetK, tetC, tetE, tetM | [127] |
| Italy | Enterococci | ampicillin, gentamicin | tetM, tetL, tetO, ermB, mef | [142] |
| Italy | Aeromonas spp., Photobacterium spp., Shewanella spp., Vibrio spp. | tetracycline, flumequine, trimethoprim | [122] | |
| Italy | Photobacterium damselae ssp. piscicida, Vibrio fluvialis, V. alginolyticus, V. parahaemolyticus, V. metschnikovii | ampicillin, carbenicillin, kanamycin, cefalothin | [143] | |
| Italy | Aeromonas spp. | ampicillin, amoxicillin, cephalothin, erythromycin, streptomycin, sulfadiazine, trimethoprim | [125] | |
| Italy | Shewanella algae, Vibrio spp. | beta-lactams, quinolones, tetracyclines, macrolides, polymyxins, chloramphenicol, fosfomycin, erythromycin | blaOXA-55-like, blaAmpC, mexB-OprM, mdtG, mdlB,tet34, tet35, tetR, eptA, cat, mdtL |
[46] |
| Lebanon | Streptococcus pneumoniae | polymyxins, chloramphenicol, fosfomycin, erythromycin | [144] | |
| Spain | Aeromonas spp., Salmonella spp., Vibrio mimicus, V. furnissii. | oxytetracycline, nitrofurantoin, oxacillin, sulfomethoxazole/trimethoprim | [145] | |
| Spain | Flavobacterium psychrophilum | oxytetracycline, florfenicol | [146] | |
| Spain | Aeromonas salmonicida | nalidixic acid, oxytetracycline | [147] | |
| Tunisia | Vibrio alginolyticus | ampicillin, erythromycin, kanamycin, cefataxime, streptomycin, trimetoprim | [148] | |
| Tunisia | Escherichia coli | tertracycline, streptomycin, ampicillin, trimethoprim, sulfamethoxazole | tetA-tetB | [149] |
| Turkey | E. coli, coliforms, fish pathogens | sulfamethoxazole, ampicillin, sulfamethoxazole, imipenem, aztreonam | ampC, blaCTX-M1, tetA, sul2, blaTEM | [150] |
| Turkey | Y. ruckeri | floR, sulI, tetC, tetD, tetE | [151] | |
| Turkey | Y. ruckeri | erythromycin, florfenicol, sulfonamide, tetracycline, trimetophrin | ermB, ermY, floR, su/I, suffll, tetA-tetG | [152] |
| Turkey | Aeromonas media, A. rivipollensis, A. salmonicida, Bacillus pumilus, B. zhangzhouensis, Hafnia alvei, Kluyvera intermedia, Pantoea spp., Pseudomonas spp., P. protegens, Staphylococcus spp., Gammaproteobacteria, Betaproteobacteria, Enterobacteriales, Burkholderiales | sulfamethacin, sulfamerazine, erythromycin, tetracycline | [153] |
5. Conclusions
In the face of undoubted advantages in economic and social terms, aquaculture in the Mediterranean basin involves some critical issues, one of which is represented by resistance to antibiotics. A link between antibiotics utilization in animals, the emergence of resistant bacterial strains, and the transfer of resistance to human pathogens have been highlighted. New insights and new research directions could include the use of new antibiotics, for instance from marine microorganisms, to be applied in aquaculture, avoiding the utilization of the same antibiotics deputed to human therapies. As for recommendations, it is important to decrease the use of antibiotics in aquaculture, improving the care of fish farms, and promoting the hygiene of the facilities.
References
- Andersson, D.I.; Hughes, D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist. Updat. 2012, 15, 162–172.
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158.
- Bengtsson-Palme, J.; Larsson, D.J. Concentrations of antibiotics predicted to select for resistant bacteria: Pro-posed limits for environmental regulation. Environ. Int. 2016, 86, 140–149.
- Miranda, C.D.; Tello, A.; Keen, P.L. Mechanisms of antimicrobial resistance in finfish aquaculture environments. Front. Microbiol. 2013, 4, 233.
- Zhu, Y.-G.; Zhao, Y.; Li, B.; Huang, C.-L.; Zhang, S.-Y.; Si-Yu, Z.; Chen, Y.-S.; Zhang, T.; Gillings, M.R.; Su, J.-Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270.
- Lin, J.; Nishino, K.; Roberts, M.C.; Tolmasky, M.; Aminov, R.I.; Zhang, L. Mechanisms of antibiotic resistance. Front. Microbiol. 2015, 6, 34.
- Romero, J.; Gloria, C.; Navarrete, P. Antibiotics in Aquaculture—Use, Abuse and Alternatives. In Health and Environment in Aquaculture; IntechOpen: Rijeka, Croatia, 2012.
- Rigos, G.; Troisi, G.M. Antibacterial Agents in Mediterranean Finfish Farming: A Synopsis of Drug Pharmacoki-netics in Important Euryhaline Fish Species and Possible Environmental Implications. Rev. Fish Biol. Fish. 2005, 15, 53–73.
- Rigos, G.; Kogiannou, D.; Padrós, F.; Cristòfol, C.; Florio, D.; Fioravanti, M.; Zarza, C. Best therapeutic practices for the use of antibacterial agents in finfish aquaculture: A particular view on European seabass (Dicentrarchus labrax) and gilthead seabream (Sparus aurata) in Mediterranean aquaculture. Rev. Aquac. 2020, 1, 39.
- FAO. The State of World Fishery and Aquaculture 2020 (SOFIA); Food and Agriculture Organization of the United Nations: Rome, Italy, 2020.
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Mar. Drugs 2017, 15, 158.
- Zago, V.; Veschetti, L.; Patuzzo, C.; Malerba, G.; Lleo, M.M. Shewanella algae and Vibrio spp. strains isolated in Italian aquaculture farms are reservoirs of antibiotic resistant genes that might constitute a risk for human health. Mar. Pollut. Bull. 2020, 154, 111057.
- Anastasiou, T.I.; Mandalakis, M.; Krigas, N.; Vézignol, T.; Lazari, D.; Katharios, P.; Dailianis, T.; Antonopoulou, E. Comparative Evaluation of Essential Oils from Medicinal-Aromatic Plants of Greece: Chemical Composition, Antioxidant Capacity and Antimicrobial Activity against Bacterial Fish Pathogens. Molecules 2019, 25, 148.
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478.
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942.
- Erauso, G.; Lakhal, F.; Bidault-Toffin, A.; Le Chevalier, P.; Bouloc, P.; Paillard, C.; Jacq, A. Evidence for the Role of Horizontal Transfer in Generating pVT1, a Large Mosaic Conjugative Plasmid from the Clam Pathogen, Vibrio tapetis. PLoS ONE 2011, 6, e16759.
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, 031.
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence eDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827.
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96.
- Yuan, K.; Wang, X.; Chen, X.; Zhao, Z.; Fang, L.; Chen, B.; Jiang, J.; Luan, T.; Chen, B. Occurrence of antibiotic re-sistance genes in extracellular and intracellular DNA from sediments collected from two types of aquaculture farms. Chemosphere 2019, 234, 520–527.
- Nielsen, K.M.; Johnsen, P.J.; Bensasson, D.; Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007, 6, 37–53.
- Poly, F.; Chenu, C.; Simonet, P.; Rouiller, J.; Monrozier, L.J. Differences between Linear Chromosomal and Supercoiled Plasmid DNA in Their Mechanisms and Extent of Adsorption on Clay Minerals. Langmuir 2000, 16, 1233–1238.
- Zarei-Baygi, A.; Smith, A.L. Intracellular versus extracellular antibiotic resistance genes in the environment: Prevalence, horizontal transfer, and mitigation strategies. Bioresour. Technol. 2021, 319, 124181.
- Liu, S.-S.; Qu, H.-M.; Yang, D.; Hu, H.; Liu, W.-L.; Qiu, Z.-G.; Hou, A.-M.; Guo, J.; Li, J.-W.; Shen, Z.-Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136.
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Genet. 2005, 3, 711–721.
- Von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173.
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting Plasmids to Limit Acquisition and Transmission of Antimicrobial Resistance. Front. Microbiol. 2020, 11, 761.
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plas-mid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1–10.
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610.
- Chandrasekaran, S.; Venkatesh, B.; Lalithakumari, D. Transfer and expression of a multiple antibiotic resistance plasmid in marine bacteria. Curr. Microbiol. 1998, 37, 347–351.
- Bergman, J.; Fineran, P.; Petty, N.; Salmond, G. Transduction: The transfer of host DNA by bacteriophages. In Encyclopedia of Microbiology, 4th ed.; Elsevier: Cambridge, MA, USA, 2019; pp. 458–473.
- Jiang, S.; Paul, J.H. Gene Transfer by Transduction in the Marine Environment. Appl. Environ. Microbiol. 1998, 64, 2780–2787.
- Brown-Jaque, M.; Calero-Cáceres, W.; Muniesa, M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 2015, 79, 1–7.
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecologi-cal network of the phage metagenome. Nature 2013, 499, 219–222.
- Landén, R.; Heierson, A.; Boman, H.G. A phage for generalized transduction in Bacillus thuringiensis and map-ping of four genes for antibiotic resistance. Microbiology 1981, 123, 49–59.
- Varga, M.; Kuntová, L.; Pantůček, R.; Mašlaňová, I.; Růžičková, V.; Doškař, J. Efficient transfer of antibiotic re-sistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 2012, 332, 146–152.
- Volkova, V.V.; Lu, Z.; Besser, T.; Gröhn, Y.T. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: Estimating the contribution of transduction to antimicrobial gene spread. Appl. Environ. Microbiol. 2014, 80, 4350–4362.
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21.
- Vlassov, V.V.; Laktionov, P.P.; Rykova, E.Y. Extracellular nucleic acids. BioEssays 2007, 29, 654–667.
- Pietramellara, G.; Ascher-Jenull, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fertil. Soils 2009, 45, 219–235.
- Stewart, G.J.; Sinigalliano, C.D. Detection of Horizontal Gene Transfer by Natural Transformation in Native and Introduced Species of Bacteria in Marine and Synthetic Sediments. Appl. Environ. Microbiol. 1990, 56, 1818–1824.
- Dubnau, D. DNA Uptake in Bacteria. Annu. Rev. Microbiol. 1999, 53, 217–244.
- Johnsborg, O.; Eldholm, V.; Håvarstein, L.S. Natural genetic transformation: Prevalence, mechanisms and function. Res. Microbiol. 2007, 158, 767–778.
- Palmen, R.; Hellingwerf, K.J. Uptake and processing of DNA by Acinetobacter calcoaceticus—A review. Gene 1997, 192, 179–190.
- Zawadzki, P.; Cohan, F.M. The size and continuity of DNA segments integrated in Bacillus transformation. Genetics 1995, 141, 1231–1243.
- Tello, A.; Austin, B.; Telfer, T. Selective Pressure of Antibiotic Pollution on Bacteria of Importance to Public Health. Environ. Health Perspect. 2012, 120, 1100–1106.
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739.
- Cattoir, V.; Nordmann, P. Plasmid-mediated quinolone resistance in gram-negative bacterial species: An update. Curr. Med. Chem. 2009, 16, 1028–1046.
- Cattoir, V.; Poirel, L.; Aubert, C.; Soussy, C.-J.; Nordmann, P. Unexpected Occurrence of Plasmid-mediated Quinolone Resistance Determinants in Environmental Aeromonas spp. Emerg. Infect. Dis. 2008, 14, 231.
- Tomova, A.; Ivanova, L.; Buschmann, A.H.; Godfrey, H.P.; Cabello, F.C. Plasmid-Mediated Quinolone Resistance (PMQR) Genes and Class 1 Integrons in Quinolone-Resistant Marine Bacteria and Clinical Isolates of Escherichia coli from an Aquacultural Area. Microb. Ecol. 2017, 75, 104–112.
- Yang, H.; Duan, G.; Zhu, J.; Zhang, W.; Xi, Y.; Fan, Q. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int. J. Antimicrob. Agents 2013, 42, 173–177.
- Poirel, L.; Cattoir, V.; Nordmann, P. Plasmid-Mediated Quinolone Resistance; Interactions between Human, Animal, and Environmental Ecologies. Front. Microbiol. 2012, 3, 24.
- Eftekhar, F.; Seyedpour, S.M. Prevalence of qnr and aac(6′)-Ib-cr Genes in Clinical Isolates of Klebsiella pneumoni-ae from Imam Hussein Hospital in Tehran. Iran. J. Med. Sci. 2015, 40, 6.
- Khajanchi, B.K.; Fadl, A.A.; Borchardt, M.A.; Berg, R.L.; Horneman, A.J.; Stemper, M.E.; Joseph, S.W.; Moyer, N.P.; Sha, J.; Chopra, A.K. Distribution of Virulence Factors and Molecular Fingerprinting of Aeromonas Species Isolates from Water and Clinical Samples: Suggestive Evidence of Water-to-Human Transmission. Appl. Environ. Microbiol. 2010, 76, 2313–2325.
- Sánchez-Céspedes, J.; Blasco, M.D.; Marti, S.; Alcalde, E.; Vila, J.; Alba, V.; Esteve, C. Plasmid-Mediated QnrS2 Determinant from a Clinical Aeromonas veronii Isolate. Antimicrob. Agents Chemother. 2008, 52, 2990–2991.
- Lavilla, S.; Gonzalez-Lopez, J.J.; Sabate, M.; Garcia-Fernandez, A.; Larrosa, M.N.; Bartolome, R.M.; Carattoli, A.; Prats, G. Prevalence of qnr genes among extended-spectrum β-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 2008, 61, 291–295.
- Park, Y.J.; Yu, J.K.; Lee, S.; Oh, E.J.; Woo, G.J. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007, 60, 868–871.
- Heuer, O.E.; Kruse, H.; Grave, K.; Collignon, P.; Karunasagar, I.; Angulo, F.J. Human Health Consequences of Use of Antimicrobial Agents in Aquaculture. Clin. Infect. Dis. 2009, 49, 1248–1253.
- Wen, Y.; Pu, X.; Zheng, W.; Hu, G. High Prevalence of Plasmid-Mediated Quinolone Resistance and IncQ Plasmids Carrying qnrS2 Gene in Bacteria from Rivers near Hospitals and Aquaculture in China. PLoS ONE 2016, 11, e0159418.
- Poirel, L.; Rodriguez-Martinez, J.M.; Mammeri, H.; Liard, A.; Nordmann, P. Origin of plasmid-mediated quino-lone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005, 49, 3523–3525.
- Cattoir, V.; Poirel, L.; Mazel, D.; Soussy, C.J.; Nordmann, P. Vibrio splendidus as the source of plasmid-mediated QnrS-Like quinolone resistance determinants. Antimicrob. Agents Chemother. 2007, 51, 2650–2651.
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014; WHO: Geneva, Switzerland, 2014; Available online: (accessed on 11 February 2021).
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260.
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial Resistance to Tetracycline: Mechanisms, Transfer, and Clinical Significance. Clin. Microbiol. Rev. 1992, 5, 387–399.
- Furushita, M.; Shiba, T.; Maeda, T.; Yahata, M.; Kaneoka, A.; Takahashi, Y.; Torii, K.; Hasegawa, T.; Ohta, M. Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl. Environ. Microbiol. 2003, 69, 5336–5342.
- Sørum, H. Antimicrobial drug resistance in fish pathogens. In Antimicrobial Resistance in Bacteria of Animal Origin; Aarestrup, F.M., Ed.; ASM Press: Washington, DC, USA, 2006; pp. 213–238.
- Rhodes, G.; Huys, G.; Swings, J.; McGann, P.; Hiney, M.; Smith, P. Distribution of oxytetracycline re-sistance plasmids between Aeromonads in hospital and aquaculture environments: Implication of Tn1721 in dis-semination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol. 2000, 66, 3883–3890.
- Briggs, C.E.; Fratamico, P.M. Molecular Characterization of an Antibiotic Resistance Gene Cluster of Salmonella typhimuriumDT104. Antimicrob. Agents Chemother. 1999, 43, 846–849.
- Angulo, F.J.; Johnson, K.R.; Tauxe, R.V.; Cohen, M.L. Origins and consequences of antimicrobial-resistant non-typhoidal Salmonella: Implications for the use of fluoroquinolones in food animals. Microb. Drug Resist. 2000, 6, 77–183.
- Angulo, F.J.; Griffin, P.M. Changes in Antimicrobial Resistance in Salmonella enterica Serovar Typhimurium. Emerg. Infect. Dis. 2000, 6, 436–437.
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133.
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143.
- Sousa, M.; Torres, C.; Barros, J.; Somalo, S.; Igrejas, G.; Poeta, P. Gilthead Seabream (Sparus aurata) as Carriers of SHV-12 and TEM-52 Extended-Spectrum Beta-Lactamases-Containing Escherichia coli Isolates. Foodborne Pathog. Dis. 2011, 8, 1139–1141.
- Eckburg, P.B.; Lister, T.; Walpole, S.; Keutzer, T.; Utley, L.; Tomayko, J.; Kopp, E.; Farinola, N.; Coleman, S. Safety, Tolerability, Pharmacokinetics, and Drug Interaction Potential of SPR741, an Intravenous Potentiator, after Single and Multiple Ascending Doses and When Combined with β-Lactam Antibiotics in Healthy Subjects. Antimicrob. Agents Chemother. 2019, 63, e00892-19.
- Ibrahim, M.E.; Abbas, M.; Al-Shahrai, A.M.; Elamin, B.K. Phenotypic Characterization and Antibiotic Re-sistance Patterns of Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Gram-Negative Bacteria in a Referral Hospital, Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2019, 6054694.
- Liu, Y.-Y.; Wang, Y.; Walsh, T.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168.
- Telke, A.A.; Rolain, J.-M. Functional genomics to discover antibiotic resistance genes: The paradigm of resistance to colistin mediated by ethanolamine phosphotransferase in Shewanella algae MARS 14. Int. J. Antimicrob. Agents 2015, 46, 648–652.
- Tomova, A.; Ivanova, L.; Buschmann, A.H.; Rioseco, M.L.; Kalsi, R.K.; Godfrey, H.P.; Cabello, F.C. Antimicrobial resistance genes in marine bacteria and human uropathogenic Escherichia coli from a region of intensive aquaculture. Environ. Microbiol. Rep. 2015, 7, 803–809.
- Bills, G.F.; Gloer, J.B. Biologically active secondary metabolites from the fungi. Fungal Kingd. 2017, 1087–1119.
- Bolton, L.F.; Kelley, L.C.; Lee, M.D.; Fedorka-Cray, P.J.; Maurer, J.J. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloram-phenicol. J. Clin. Microbiol. 1999, 37, 1348–1351.
- Boyd, D.; Peters, G.A.; Cloeckaert, A.; Boumedine, K.S.; Chaslus-Dancla, E.; Imberechts, H.; Mulvey, M.R. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 2001, 183, 5725–5732.
- Clark, G.M.; Kaufmann, A.F.; Gangarosa, E.J. Epidemiology of an international outbreak of Salmonella Agona. Lancet 1973, 2, 490–493.
- Weber, J.T.; Mintz, E.D.; Canizares, R.; Semiglia, A.; Gomez, I.; Sempertegui, R.; Davila, A.; Greene, K.D.; Puhr, N.D.; Cameron, D.N.; et al. Epidemic cholera in Ecuador: Multidrug-resistance and transmission by water and seafood. Epidemiol. Infect. 1994, 112, 1–11.
- Nogales, B.; Lanfranconi, M.P.; Piña-Villalonga, J.M.; Bosch, R. Anthropogenic perturbations in marine microbial communities. FEMS Microbiol. Rev. 2011, 35, 275–298.
- Pridgeon, J.W.; Klesius, P.H. Major bacterial diseases in aquaculture and their vaccine development. Cab Rev. 2012, 7, 1–16.
- Chelossi, E.; Vezzulli, L.; Milano, A.; Branzoni, M.; Fabiano, M.; Riccardi, G.; Banat, I.M. Antibiotic resistance of benthic bacteria in fish-farm and control sediments of the Western Mediterranean. Aquaculture 2003, 219, 83–97.
- Bakopoulos, V.; Adams, A.; Richards, R.H. Some biochemical properties and antibiotic sensitivities of Pasteurel-la piscicida isolated in Greece and comparison with strains from Japan, France and Italy. J. Fish Dis. 1995, 18, 1–7.
- Zorilla, I.; Chabrillon, M.; Arijo, S.; Diaz-Rosales, P.; Martinez-Manzanares, E.; Babelona, M.; Morinigo, M. Bacteria recovered from diseased cultured gilthead seabream (Sparus aurata) in southwestern Spain. Aquaculture 2003, 218, 11–20.
- Smith, P.; Christofilogiannis, P. Application of Normalised Resistance Interpretation to the detection of multiple low-level resistance in strains of Vibrio anguillarum obtained from Greek fish farms. Aquaculture 2007, 272, 223–230.
- Labella, A.; Gennari, M.; Ghidini, V.; Trento, I.; Manfrin, A.; Borrego, J.; Lleo, M. High incidence of antibiotic multi-resistant bacteria in coastal areas dedicated to fish farming. Mar. Pollut. Bull. 2013, 70, 197–203.
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134.
- Ottaviani, D.; Bacchiocchi, I.; Masini, L.; Francesca, L.; Carraturo, A.; Giammarioli, M.; Sbaraglia, G. Antimicrobial susceptibility of potentially pathogenic halophilic vibrios isolated from seafood. Int. J. Antimicrob. Agents 2001, 18, 135–140.
- Scarano, C.; Piras, F.; Virdis, S.; Ziino, G.; Nuvoloni, R.; Dalmasso, A.; De Santis, E.; Spanu, C. Antibiotic resistance of Aeromonas ssp. strains isolated from Sparus aurata reared in Italian mariculture farms. Int. J. Food Microbiol. 2018, 284, 91–97.
- Salgueiro, V.; Manageiro, V.; Bandarra, N.M.; Reis, L.; Ferreira, E.; Caniça, M. Bacterial Diversity and Antibiotic Susceptibility of Sparus aurata from Aquaculture. Microorganisms 2020, 8, 1343.
- Nikolakopoulou, T.L.; Giannoutsou, E.P.; Karabatsou, A.A.; Karagouni, A.D. Prevalence of tetracycline re-sistance genes in Greek seawater habitats. J. Microbiol. 2008, 46, 633.
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Genet. 2010, 8, 251–259.
- Sutili, F.J.; Gressler, L.T. Chapter 3—Antimicrobial agents. In Aquaculture Pharmacology, 1st ed.; Kibenge, F.S.B., Baldisserotto, B., Chong, R.S.-M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 131–196.
- Cassini, A. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66.
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.-C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; De Verdal, H.; Gozlan, R.E. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020, 11, 1–8.
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.B.; Levin, S.A.S.; Bonhoeffer, S.; Laxminarayan, R.R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352.
More
Information
Subjects:
Environmental Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revision:
1 time
(View History)
Update Date:
08 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




