Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Changbo Qu | + 1734 word(s) | 1734 | 2021-06-02 10:25:14 | | | |
| 2 | Vicky Zhou | -2 word(s) | 1732 | 2021-06-03 03:40:13 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Qu, C. Histamine H1 Receptor Antagonists. Encyclopedia. Available online: https://encyclopedia.pub/entry/10452 (accessed on 08 February 2026).
Qu C. Histamine H1 Receptor Antagonists. Encyclopedia. Available at: https://encyclopedia.pub/entry/10452. Accessed February 08, 2026.
Qu, Changbo. "Histamine H1 Receptor Antagonists" Encyclopedia, https://encyclopedia.pub/entry/10452 (accessed February 08, 2026).
Qu, C. (2021, June 03). Histamine H1 Receptor Antagonists. In Encyclopedia. https://encyclopedia.pub/entry/10452
Qu, Changbo. "Histamine H1 Receptor Antagonists." Encyclopedia. Web. 03 June, 2021.
Copy Citation
Histamine H1 receptor (H1 receptor) antagonists are widely prescribed medications to treat allergic diseases, while recently it has emerged that they show significant promise as anti-SARS-CoV-2 agents.
COVID-19
NF-κB signaling
H1 receptor antagonists
treatment
drugs
1. Introduction
Coronavirus disease 2019 (COVID-19), an emerging respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is swiftly leading to global health issues and becoming a pandemic worldwide. It forces much of the world to adopt a lockdown mode, causing enormous economic fallout and human suffering. Most patients with COVID-19 are either asymptomatic or show mild symptoms; however in some cases, patients progress to severe lung injuries and eventually develop multiple organ failure [1][2].
SARS-CoV-2 is a single-stranded, positive-sense RNA virus (++ssRNA) [3]. The SARS-CoV-2 genome possesses an 82% sequence identity to that of SARS-CoV and MERS-CoV. Four structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins have been identified in SARS-CoV-2. These protein sequences are also highly similar to that of SARS-CoV and MERS-CoV [4]. The viral structural proteins play vital roles in determining the viral life cycle, and thus provide potential therapeutic targets [5]. SARS-CoV-2 engages SARS-CoV angiotensin converting enzyme 2 (ACE2) receptor for entry and transmembrane serine protease (TMPRSS2) for S protein priming. After entering the cell, SARS-CoV-2 is subsequently taken up into endosomes and then fused with lysosomal membranes. Eventually, SARS-CoV-2 virions are released from the cell through exocytosis (Figure 1) [6]. SARS-CoV-2 infection can cause severe respiratory pathologies and lung injuries [7]. The severity of the lung injuries is correlated with the production of a cytokine storm by the macrophages during SARS-CoV-2 infection. High levels of cytokines including IL-2, IL-10, GCSF, IP-10, MCP-1, IL-7, TNF-α, and MIP-1A were observed in COVID-19 patients at high risk of mortality [1]. In parallel, an enhanced concentration of perivascular and septal mast cells was found in post-mortem lung biopsies of COVID-19 [8]. Mast cells synthesize and secrete inflammatory mediators including histamine. The roles of mast cells in SARS-CoV-2 infection have been frequently discussed [9][10][11][12]. Whether histamine released by mast cell activation during SARS-CoV-2 infection contributes to the severity of lung injury remains to be elucidated [13][14].
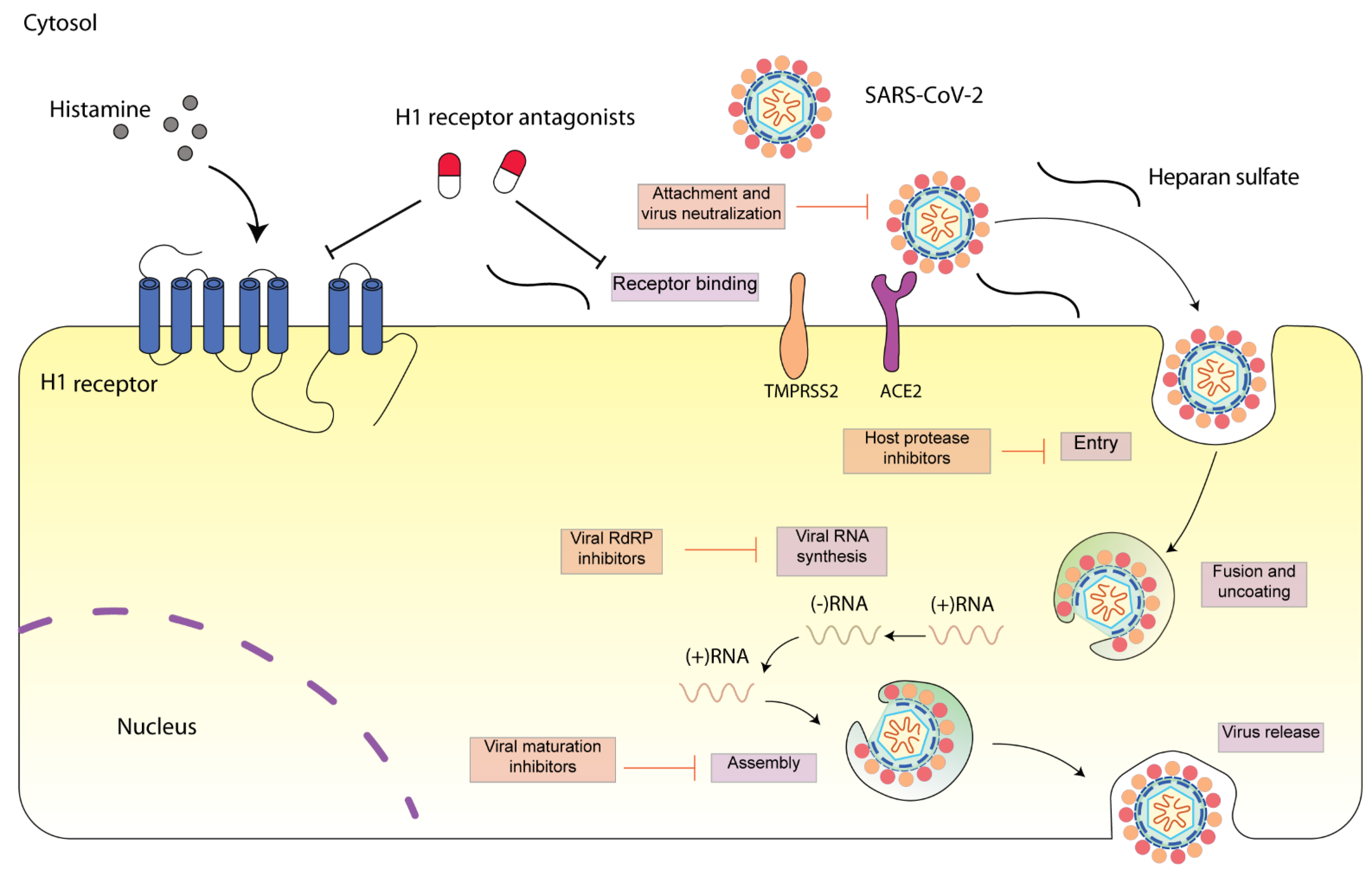 Figure 1. Schematic diagram presenting life cycle of SARS-CoV-2 and relevant inhibitors. SARS-CoV-2 cell entry begins with binding of the spike S protein to ACE2, a process that is facilitated by TMPRSS2. SARS-CoV-2 enters the cell through endocytosis, and then the virus is uncoated in the acidic environment of lysosomes. After that, SARS-CoV-2 RNA is released, followed by the reproduction of virus genome and viral proteins. Then, the viral components are assembled and released via exocytosis [15]. Each step can be targeted by relevant inhibitors. H1 receptor antagonists may inhibit SARS-CoV-2 either via H1 receptor or via ACE2 receptor. SARS-CoV-2 spike protein interacts with both cellular heparan sulfate and ACE2 through its receptor-binding domain (RBD) [16]. H1 receptor antagonists may disrupt the interaction between heparan sulfate and spike protein, inhibiting SARS-CoV-2 entry.
Figure 1. Schematic diagram presenting life cycle of SARS-CoV-2 and relevant inhibitors. SARS-CoV-2 cell entry begins with binding of the spike S protein to ACE2, a process that is facilitated by TMPRSS2. SARS-CoV-2 enters the cell through endocytosis, and then the virus is uncoated in the acidic environment of lysosomes. After that, SARS-CoV-2 RNA is released, followed by the reproduction of virus genome and viral proteins. Then, the viral components are assembled and released via exocytosis [15]. Each step can be targeted by relevant inhibitors. H1 receptor antagonists may inhibit SARS-CoV-2 either via H1 receptor or via ACE2 receptor. SARS-CoV-2 spike protein interacts with both cellular heparan sulfate and ACE2 through its receptor-binding domain (RBD) [16]. H1 receptor antagonists may disrupt the interaction between heparan sulfate and spike protein, inhibiting SARS-CoV-2 entry.In most cases, the excess lung inflammation response caused by SARS-CoV-2 is self-competent; however, in some patients, it is unbalanced and non-competent, with age and comorbidities such as arterial hypertension or diabetes being acknowledged as risk factors. As a consequence, these patients require hospitalization and need to be managed appropriately. Considering the alleviation of the inflammatory response and concomitant lung injuries, anti-inflammatory drugs (non-steroidal anti-inflammatory drugs (NSAIDs) or corticosteroids) are being administered to COVID-19 patients with various treatment regimens [17][18]. However, debates exist regarding their clinical use in COVID-19 patients [19][20]. For instance, ibuprofen, an over-the-counter medication used for the treatment of pain and fever in COVID-19, has been found to increase ACE2 levels [21]. In terms of corticosteroids, a recent study showed that low-dose dexamethasone, particularly in critically ill COVID-19 patients (i.e., ICU-hospitalized patients with respiratory distress), significantly improved patient survival [22]. Nevertheless, it may disrupt the immunocompetence in COVID-19 patients [23][24][25].
Histamine and its receptors play an important role in the progression of various allergic diseases [26]. Notably, the histamine H1 receptor (H1 receptor) has been reported to regulate allergic lung responses; therefore, its antagonists have been used to treat airway inflammation [27]. Beyond its role in mediating airway inflammation, our recent experimental work has identified that deptropine, a classical H1 receptor antagonist used to treat asthmatic symptoms, potently inhibits hepatitis E virus replication [28]. Along with our finding, a growing body of evidence also demonstrated that H1 receptor antagonists can inhibit various RNA virus infections [29][30].
2. Potential Anti-SARS-CoV-2 Mechanisms of H1 Receptor Antagonists
Preclinical data suggest that H1 receptor antagonists limit viral entry of Ebola/Marburg [31] or Hepatitis C virus (HCV) [32] even if the molecular pathways explaining these antiviral effects remain largely obscure. For example, diphenyl-piperazines (e.g., chlorcyclizine, cyclizine, and hydroxyzine), cycloheptene-piperidines (e.g., cyproheptadine, ketotifen, loratadine, and desloratadine), and phenothiazines (e.g., mequitazine and trimeprazine) have been found to inhibit HCV cell entry. Interestingly, the potential anti-HCV mechanisms of these drugs are likely independent of H1 receptor [33].
2.1. Intervention of Early Step of SARS-CoV-2 Infection
Consistent with HCV and Ebola, the H1 receptor antagonists may also inhibit SARS-CoV-2 infection via intervention in the early step of virus replication. It may prevent SARS-CoV-2 from entering the cell host through binding ACE2 [34]. Moreover, evidence showed that H1 receptor antagonists could inhibit SARS-CoV-2 infection through binding sigma receptor-1 (for hydroxyzine and diphenhydramine), an endoplasmic reticulum resident chaperone protein participating in early steps of SARS-CoV-2 replication [35][34]. Heparin belongs to glycosaminoglycan molecules which include heparan sulphate. During virus entry, SARS-CoV-2 spike protein was demonstrated to interact with both cellular heparan sulfate and ACE2 [16]. Therefore, disrupting the interaction between heparan sulphate and spike protein by exogenous and competitive heparin mimetics could limit the SARS-CoV-2 virus entering the cell, resulting in suppression of the inflammatory responses [36]. Histamine has been shown capable of binding to heparan sulfate [37][38]. It is interesting to investigate the involvement of heparan sulfate in the H1 receptor antagonist-mediated anti-SARS-CoV-2 activity.
Unlike H1 receptor, the proposed anti-SARS-CoV-2 mechanisms for H2 receptor antagonists are inhibition of Type 2 transmembrane serine protease (TMPRSS2) and 3-chymotrypsin-like protease (3CLpro) by famotidine [39]. However, a recent study revealed that the principal mechanism of action of famotidine for relieving COVID-19 symptoms is attributed to its on-target effect on H2 receptor activity, and is likely independent of other histamine receptors determined by competition assay [14]. Most recently, molecular docking results suggest that famotidine and cimetidine inhibit SARS-CoV-2 replication through the inhibition of non-structural proteins, including NSP3, NSP7/8 complex, and NSP9 [40]. The modes of action of anti-SARS-CoV-2 activity of famotidine needs to be further clarified.
2.2. NF-κB-Mediated Antiviral Activity by H1 Receptor Antagonist
Activation of cellular inflammation was observed following histamine binding to H1 receptor. This activation was mediated by phospholipase C (PLC) [41], protein kinase C (PKC), and NF-κB signaling pathways [42]. Our recent experimental work demonstrated that deptropine could potently inhibit HEV replication. The anti-HEV effect of deptropine requires the inhibition of NF-κB activity and is likely dispensable of H1 receptor [28]. Interestingly, activation of NF-κB in lung epithelial cells was found to facilitate SARS-CoV-2 propagation, and H1 receptor antagonists were able to suppress the activation of NF-κB in lung epithelial cells [43][44]. The above evidence together prompts us to hypothesize that the potential mechanism by which H1 receptor antagonists inhibit SARS-CoV-2 may include their inhibitory effects on NF-κB [28]. Of note, H1 receptor antagonists can inhibit NF-κB through H1 receptor-dependent and -independent mechanisms [44]. The implication of H1 receptor and the NF-κB signaling pathway in a H1 receptor antagonists-mediated anti-SARS-CoV-2 effect is intriguing and needs to be further explored.
2.3. Other Potential Antiviral Mechanisms
The observation that H1 receptor antagonists exert antiviral effects, however, also fits well with recent observations that the action of H1 receptor in physiology is much broader than the conventional view that such action is mainly restricted to allergic inflammation. The endogenous histamine stimulates oleoylethanolamide biosynthesis in the liver via H1 receptor activation, regulating liver ketogenesis [45]. Epidemiological evidence as well as preclinical studies indicate that H1 receptor antagonists are associated with increased food intake, body weight, and obesity [46]. The H1 receptor was also shown to play a vital role in the control of cellular metabolism. Genetic deletion of H1 receptor in mice leads to increased abdominal adiposity and glucose intolerance [47]. Metabolism and immune responses are two fundamental biological processes that protect the cell host from viral infection. Interestingly, it has been proposed that H1 receptor can modulate innate immunity beyond its role in regulating allergic reactions [48]. Thus, the blockage of H1 receptor by its antagonists may affect the SARS-CoV-2 replication via mediation of the crosstalk between metabolism and immune responses.
3. Opportunities and Challenges
There are various advantages in the application of H1 receptor antagonists for the treatment of COVID-19. First, these drugs are relatively inexpensive and readily to be used. Second, although the chronic inflammation may not be the primary driver of pathology, using H1 receptor antagonists can improve patient outcomes due to its natural role in reducing inflammation. However, it also should be noted that the first-generation H1 receptor antagonists are not strictly selective for the H1 receptor. They contribute to a spectrum of dopaminergic and cholinergic reactions, resulting in significant adverse effects in the central nervous system due to their capability of penetrating the blood-brain barrier [49]. Moreover, they are able to regulate cardiac channels, leading to wide range of side effects, including sedation, insomnia, and hyperactivity [50]. Besides that, H1 receptor antagonists have been associated with an increase in seizure susceptibility [51]; therefore, H1 receptor antagonists in most cases are not recommended to be prescribed to COVID-19 patients with epilepsy or febrile seizures. Another factor that may hinder the clinical use of H1 receptor antagonists in COVID-19 is the impairment of innate immune responses by these drugs [52].
As mentioned above, both loratadine and desloratadine could inhibit SARS-CoV-2 infection. They are new-generation H1 receptor antagonists which are highly selective to H1 receptor with less central nervous system side effects. Besides their role in reducing lung inflammation induced by histamine, they can suppress a number of other inflammatory activities [53], which make them ideal medications for relieving the excess inflammatory response of COVID-19. Overall, with studies describing H1 receptor antagonists to be capable of treating viral diseases, recent findings take us one step closer to understanding the underlying antiviral mechanism and the advantages of the H1 receptor antagonists in the treatment of COVID-19. In the future, the use of H1 receptor antagonists as antiviral agents will further advance our understanding of how these agents work beyond their intrinsic effects on the histamine receptor, and bear potential implications for the real-world treatment of COVID-19 patients.
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422.
- Chan, J.F.; To, K.K.; Tse, H.; Jin, D.Y.; Yuen, K.Y. Interspecies transmission and emergence of novel viruses: Lessons from bats and birds. Trends Microbiol. 2013, 21, 544–555.
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878.
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021, 12, 961.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8.
- Aggarwal, N.R.; King, L.S.; D’Alessio, F.R. Diverse macrophage populations mediate acute lung inflammation and resolution. Am. J. Physiol. Cell. Mol. Physiol. 2014, 306, L709–L725.
- Junior, J.D.S.M.; Miggiolaro, A.F.R.D.S.; Nagashima, S.; De Paula, C.B.V.; Baena, C.P.; Scharfstein, J.; De Noronha, L. Mast Cells in Alveolar Septa of COVID-19 Patients: A Pathogenic Pathway That May Link Interstitial Edema to Immunothrombosis. Front. Immunol. 2020, 11, 574862.
- Kritas, S.K.; Ronconi, G.; Caraffa, A.; E Gallenga, C.; Ross, R.; Conti, P. Mast cells contribute to coronavirus-induced inflammation: New anti-inflammatory strategy. J. Biol. Regul. Homeost. Agents 2020, 34, 9–14.
- Kilinc, E.; Kilinc, Y.B. Mast cell stabilizers as a supportive therapy can contribute to alleviate fatal inflammatory responses and severity of pulmonary complications in COVID-19 infection. Anatol. Clin. J. Med. Sci. 2020, 25, 111–118.
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e2.
- Theoharides, T.C. COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. BioFactors 2020, 46, 306–308.
- Conti, P.; Caraffa, A.; Tetè, G.; Gallenga, C.E.; Ross, R.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Ronconi, G. Mast cells activated by SARS-CoV-2 release histamine which increases IL-1 levels causing cytokine storm and inflammatory reaction in COVID-19. J. Biol. Regul. Homeost Agents 2020, 34, 1629–1632.
- Malone, R.W.; Tisdall, P.; Fremont-Smith, P.; Liu, Y.; Huang, X.-P.; White, K.M.; Miorin, L.; Moreno, E.; Alon, A.; Delaforge, E.; et al. COVID-19: Famotidine, Histamine, Mast Cells, and Mechanisms. Front. Pharmacol. 2021, 12, 633680.
- Murgolo, N.; Therien, A.G.; Howell, B.; Klein, D.; Koeplinger, K.; Lieberman, L.A.; Adam, G.C.; Flynn, J.; McKenna, P.; Swaminathan, G.; et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathog. 2021, 17, e1009225.
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 2020, 183, 1043–1057.e15.
- Galimberti, F.; McBride, J.; Cronin, M.; Li, Y.; Fox, J.; Abrouk, M.; Herbst, A.; Kirsner, R.S. Evidence-based best practice advice for patients treated with systemic immunosuppressants in relation to COVID-19. Clin. Dermatol. 2020, 38, 775–780.
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613.
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; El Jammal, T.; Walzer, T.; François, B.; Sève, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567.
- Russell, B.; Moss, C.; Rigg, A.; Van Hemelrijck, M. COVID-19 and treatment with NSAIDs and corticosteroids: Should we be limiting their use in the clinical setting? Ecancer Med. Sci. 2020, 14, 1023.
- Sridharan, G.K.; Kotagiri, R.; Chandiramani, V.H.; Mohan, B.P.; Vegunta, R.; Vegunta, R.; Rokkam, V.R.P. COVID-19 and Avoiding Ibuprofen. How Good Is the Evidence? Am. J. Ther. 2020, 27, e400–e402.
- Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; Elmahi, E.; et al. Dexamethasone in hospitalized patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704.
- McGee, S.; Hirschmann, J. Use of corticosteroids in treating infectious diseases. Arch. Intern. Med. 2008, 168, 1034–1046.
- Yang, J.-W.; Fan, L.-C.; Miao, X.-Y.; Mao, B.; Li, M.-H.; Lu, H.-W.; Liang, S.; Xu, J.-F. Corticosteroids for the treatment of human infection with influenza virus: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2015, 21, 956–963.
- Saghazadeh, A.; Rezaei, N. Towards treatment planning of COVID-19: Rationale and hypothesis for the use of multiple immunosuppressive agents: Anti-antibodies, immunoglobulins, and corticosteroids. Int. Immunopharmacol. 2020, 84, 106560.
- Thurmond, R.L.; Gelfand, E.W.; Dunford, P.J. The role of histamine H1 and H4 receptors in allergic inflammation: The search for new antihistamines. Nat. Rev. Drug Discov. 2008, 7, 41–53.
- Togias, A. H1-receptors: Localization and role in airway physiology and in immune functions. J. Allergy Clin. Immunol. 2003, 112 (Suppl. 4), S60–S68.
- Qu, C.; Li, Y.; Li, Y.; Yu, P.; Li, P.; Donkers, J.M.; van de Graaf, S.F.J.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. FDA-drug screening identifies deptropine inhibiting hepatitis E virus involving the NF-κB-RIPK1-caspase axis. Antivir. Res. 2019, 170, 104588.
- Lui, C.T. Prescription practice of antihistamines for acute upper respiratory tract infections in pediatric patients in a local emergency department in Hong Kong. World J. Emerg. Med. 2017, 8, 47–54.
- He, S.; Lin, B.; Chu, V.; Hu, Z.; Hu, X.; Xiao, J.; Wang, A.Q.; Schweitzer, C.J.; Li, Q.; Imamura, M.; et al. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci. Transl. Med. 2015, 7, 282ra49.
- Cheng, H.; Lear-Rooney, C.M.; Johansen, L.; Varhegyi, E.; Chen, Z.W.; Olinger, G.G.; Rong, L. Inhibition of Ebola and Marburg Virus Entry by G Protein-Coupled Receptor Antagonists. J. Virol. 2015, 89, 9932–9938.
- Rolt, A.; Le, D.; Hu, Z.; Wang, A.Q.; Shah, P.; Singleton, M.; Hughes, E.; E Dulcey, A.; He, S.; Imamura, M.; et al. Preclinical Pharmacological Development of Chlorcyclizine Derivatives for the Treatment of Hepatitis C Virus Infection. J. Infect. Dis. 2018, 217, 1761–1769.
- Pietschmann, T. Clinically Approved Ion Channel Inhibitors Close Gates for Hepatitis C Virus and Open Doors for Drug Repurposing in Infectious Viral Diseases. J. Virol. 2016, 91, 01914–01916.
- Reznikov, L.R.; Norris, M.H.; Vashisht, R.; Bluhm, A.P.; Li, D.; Liao, Y.-S.J.; Brown, A.; Butte, A.J.; Ostrov, D.A. Identification of antiviral antihistamines for COVID-19 repurposing. Biochem. Biophys. Res. Commun. 2021, 538, 173–179.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- Yu, M.; Zhang, T.; Zhang, W.; Sun, Q.; Li, H.; Li, J.P. Elucidating the interactions between heparin/heparan sulfate and SARS-CoV-2-related proteins-an important strategy for developing novel therapeutics for the COVID-19 pandemic. Front. Mol. Biosci. 2020, 7, 628551.
- Jooyandah, F.; Moore, J.; Davies, J.; Jooyandeh, F. The Mechanism of Histamine Binding to Heparin. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979, 35, 177–182.
- Rabenstein, D.L.; Bratt, P.; Peng, J. Quantitative Characterization of the Binding of Histamine by Heparin†. Biochemistry 1998, 37, 14121–14127.
- Loffredo, M.; Lucero, H.; Chen, D.Y.; O’Connell, A.; Bergqvist, S.; Munawar, A.; Bandara, A.; De Graef, S.; Weeks, S.D.; Douam, F.; et al. The in-vitro effect of famotidine on SARS-CoV-2 proteases and virus replication. Sci. Rep. 2021, 11, 5433.
- Ishola, A.A.; Joshi, T.; Abdulai, S.I.; Tijjani, H.; Pundir, H.; Chandra, S. Molecular basis for the repurposing of histamine H2-receptor antagonist to treat COVID-19. J. Biomol. Struct. Dyn. 2021, 1–18.
- Smit, M.J.; Hoffmann, M.; Timmerman, H.; Leurs, R. Molecular properties and signalling pathways of the histamine H1 receptor. Clin. Exp. Allergy 1999, 29, 19–28.
- Matsubara, M.; Tamura, T.; Ohmori, K.; Hasegawa, K. Histamine H1 receptor antagonist blocks histamine-induced proinflammatory cytokine production through inhibition of Ca2+-dependent protein kinase C, Raf/MEK/ERK and IKK/IκB/NF-κB signal cascades. Biochem. Pharmacol. 2005, 69, 433–449.
- Kircheis, R.; Haasbach, E.; Lueftenegger, D.; Heyken, W.T.; Ocker, M.; Planz, O. NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front Immunol. 2020, 11, 598444.
- Roumestan, C.; Henriquet, C.; Gougat, C.; Michel, A.; Bichon, F.; Portet, K.; Jaffuel, D.; Mathieu, M. Histamine H1-receptor antagonists inhibit nuclear factor-kappaB and activator protein-1 activities via H1-receptor-dependent and -independent mechanisms. Clin. Exp. Allergy 2008, 38, 947–956.
- Misto, A.; Provensi, G.; Vozella, V.; Passani, M.B.; Piomelli, D. Mast Cell-Derived Histamine Regulates Liver Ketogenesis via Oleoylethanolamide Signaling. Cell Metab. 2019, 29, 91–102.e5.
- Tabarean, I.V. Histamine receptor signaling in energy homeostasis. Neuropharmacology 2016, 106, 13–19.
- Wang, K.-Y.; Tanimoto, A.; Yamada, S.; Guo, X.; Ding, Y.; Watanabe, T.; Watanabe, T.; Kohno, K.; Hirano, K.-I.; Tsukada, H.; et al. Histamine Regulation in Glucose and Lipid Metabolism via Histamine Receptors: Model for Nonalcoholic Steatohepatitis in Mice. Am. J. Pathol. 2010, 177, 713–723.
- Anvari, E.; Wang, X.; Sandler, S.; Welsh, N. The H1-receptor antagonist cetirizine ameliorates high-fat diet-induced glucose intolerance in male C57BL/6 mice, but not diabetes outcome in female non-obese diabetic (NOD) mice. Upsala J. Med. Sci. 2015, 120, 40–46.
- Estelle, F.; Simons, R. H1-receptor antagonists: Safety issues. Ann Allergy Asthma Immunol. 1999, 83, 481–488.
- Criado, P.R.; Criado, R.F.; Maruta, C.W.; Machado Filho, C. Histamine, histamine receptors and antihistamines: New concepts. An. Bras. Dermatol. 2010, 85, 195–210.
- Takano, T.; Sakaue, Y.; Sokoda, T.; Sawai, C.; Akabori, S.; Maruo, Y.; Taga, T.; Ohno, M.; Takeuchi, Y. Seizure Susceptibility Due to Antihistamines in Febrile Seizures. Pediatr. Neurol. 2010, 42, 277–279.
- Metz, M.; Doyle, E.; Bindslev-Jensen, C.; Watanabe, T.; Zuberbier, T.; Maurer, M. Effects of Antihistamines on Innate Immune Responses to Severe Bacterial Infection in Mice. Int. Arch. Allergy Immunol. 2011, 155, 355–360.
- Hou, Y.; Ge, S.; Li, X.; Wang, C.; He, H.; He, L. Testing of the inhibitory effects of loratadine and desloratadine on SARS-CoV-2 spike pseudotyped virus viropexis. Chem. Biol. Interact 2021, 338, 109420.
More
Information
Subjects:
Pharmacology & Pharmacy
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
3.9K
Revisions:
2 times
(View History)
Update Date:
03 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




