
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Byung Cheon Lee | + 2136 word(s) | 2136 | 2021-05-27 12:09:27 | | | |
| 2 | Vivi Li | Meta information modification | 2136 | 2021-06-03 08:08:04 | | |
Video Upload Options
Selenium is a vital trace element present as selenocysteine (Sec) in proteins that are, thus, known as selenoproteins. Humans have 25 selenoproteins, most of which are functionally characterized as oxidoreductases, where the Sec residue plays a catalytic role in redox regulation and antioxidant activity. Glutathione peroxidase plays a pivotal role in scavenging and inactivating hydrogen and lipid peroxides, whereas thioredoxin reductase reduces oxidized thioredoxins as well as non-disulfide substrates, such as lipid hydroperoxides and hydrogen peroxide. Selenoprotein R protects the cell against oxidative damage by reducing methionine-R-sulfoxide back to methionine. Selenoprotein O regulates redox homeostasis with catalytic activity of protein AMPylation. Moreover, endoplasmic reticulum (ER) membrane selenoproteins (SelI, K, N, S, and Sel15) are involved in ER membrane stress regulation. Selenoproteins containing the CXXU motif (SelH, M, T, V, and W) are putative oxidoreductases that participate in various cellular processes depending on redox regulation.
1. Introduction
2. Selenocysteine in Selenoproteins
3. Selenoprotein R
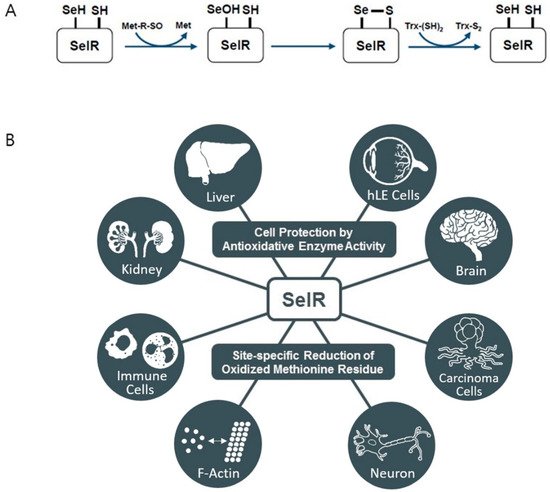
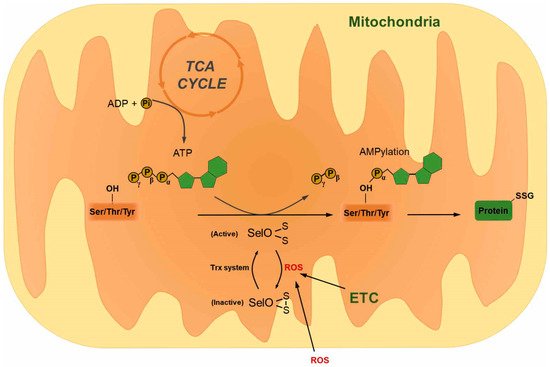
4. Selenoprotein O
References
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40.
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84.
- Dalle-Donne, I.; Rossi, R.; Colombo, R.; Giustarini, D.; Milzani, A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006, 52, 601–623.
- Dhalla, N.S.; Temsah, R.M.; Netticadan, T. Role of oxidative stress in cardiovascular diseases. J. hypertens. 2000, 18, 655–673.
- Jenner, P. Oxidative stress in Parkinson’s disease. Ann. Neurol. 2003, 53 (Suppl. 3), S26–S36; discussion S36–S38.
- Sayre, L.M.; Smith, M.A.; Perry, G. Chemistry and biochemistry of oxidative stress in neurodegenerative disease. Curr. Med. Chem. 2001, 8, 721–738.
- Brownlee, M.; Cerami, A. The biochemistry of the complications of diabetes mellitus. Annu. Rev. Biochem. 1981, 50, 385–432.
- Kasparova, S.; Brezova, V.; Valko, M.; Horecky, J.; Mlynarik, V.; Liptaj, T.; Vancova, O.; Ulicna, O.; Dobrota, D. Study of the oxidative stress in a rat model of chronic brain hypoperfusion. Neurochem. Int. 2005, 46, 601–611.
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. gerontol. 1956, 11, 298–300.
- Choi, S.M.; Kim, D.-H.; Chun, K.-S.; Choi, J.-S. Carnosol induces apoptotic cell death through ROS-dependent inactivation of STAT3 in human melanoma G361 cells. Appl. Biol. Chem. 2019, 62.
- Utaipan, T.; Boonyanuphong, P.; Chuprajob, T.; Suksamrarn, A.; Chunglok, W. A trienone analog of curcumin, 1,7-bis(3-hydroxyphenyl)-1,4,6-heptatrien-3-one, possesses ROS- and caspase-mediated apoptosis in human oral squamous cell carcinoma cells in vitro. Appl. Biol. Chem. 2020, 63.
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203.
- Wang, N.; Tan, H.Y.; Li, S.; Xu, Y.; Guo, W.; Feng, Y. Supplementation of Micronutrient Selenium in Metabolic Diseases: Its Role as an Antioxidant. Oxid. Med. Cell Longev. 2017, 2017, 7478523.
- Wessjohann, L.A.; Schneider, A.; Abbas, M.; Brandt, W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 2007, 388, 997–1006.
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. et biophy. acta 2009, 1790, 1424–1428.
- Gromer, S.; Wissing, J.; Behne, D.; Ashman, K.; Schirmer, R.H.; Flohe, L.; Becker, K. A hypothesis on the catalytic mechanism of the selenoenzyme thioredoxin reductase. Biochem. J. 1998, 332, 591–592.
- Gorlatov, S.N.; Stadtman, T.C. Human selenium-dependent thioredoxin reductase from HeLa cells: Properties of forms with differing heparin affinities. Arch. Biochem. Biophys. 1999, 369, 133–142.
- Chung, S.S.; Kim, M.; Youn, B.S.; Lee, N.S.; Park, J.W.; Lee, I.K.; Lee, Y.S.; Kim, J.B.; Cho, Y.M.; Lee, H.K.; et al. Glutathione peroxidase 3 mediates the antioxidant effect of peroxisome proliferator-activated receptor gamma in human skeletal muscle cells. Mol. Cell. Biol. 2009, 29, 20–30.
- Lee, S.R.; Bar-Noy, S.; Kwon, J.; Levine, R.L.; Stadtman, T.C.; Rhee, S.G. Mammalian thioredoxin reductase: Oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc. Natl. Acad. Sci. USA 2000, 97, 2521–2526.
- Quan, S.; Schneider, I.; Pan, J.; Von Hacht, A.; Bardwell, J.C. The CXXC motif is more than a redox rheostat. J. Biol. Chem. 2007, 282, 28823–28833.
- Johansson, L.; Gafvelin, G.; Arner, E.S. Selenocysteine in proteins-properties and biotechnological use. Biochim. biophy. Acta 2005, 1726, 1–13.
- Berry, M.J.; Banu, L.; Chen, Y.Y.; Mandel, S.J.; Kieffer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3’ untranslated region. Nature 1991, 353, 273–276.
- Bellinger, F.P.; Raman, A.V.; Reeves, M.A.; Berry, M.J. Regulation and function of selenoproteins in human disease. Biochem. J. 2009, 422, 11–22.
- Mattmiller, S.A.; Carlson, B.A.; Sordillo, L.M. Regulation of inflammation by selenium and selenoproteins: Impact on eicosanoid biosynthesis. J. Nutr. Sci. 2013, 2, e28.
- Fairweather-Tait, S.J.; Collings, R.; Hurst, R. Selenium bioavailability: Current knowledge and future research requirements. Am. J. Clin. Nutr. 2010, 91, 1484S–1491S.
- Hatfield, D.L.; Gladyshev, V.N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002, 22, 3565–3576.
- Spallholz, J.E. Selenomethionine and Methioninase: Selenium Free Radical Anticancer Activity. Methods Mol. Biol. 2019, 1866, 199–210.
- Luchman, H.A.; Villemaire, M.L.; Bismar, T.A.; Carlson, B.A.; Jirik, F.R. Prostate epithelium-specific deletion of the selenocysteine tRNA gene Trsp leads to early onset intraepithelial neoplasia. Am. J. Pathol. 2014, 184, 871–877.
- Bosl, M.R.; Takaku, K.; Oshima, M.; Nishimura, S.; Taketo, M.M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl. Acad. Sci. USA 1997, 94, 5531–5534.
- Kumaraswamy, E.; Carlson, B.A.; Morgan, F.; Miyoshi, K.; Robinson, G.W.; Su, D.; Wang, S.; Southon, E.; Tessarollo, L.; Lee, B.J.; et al. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol. Cell. Biol. 2003, 23, 1477–1488.
- Papp, L.V.; Lu, J.; Holmgren, A.; Khanna, K.K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007, 9, 775–806.
- Burk, R.F.; Hill, K.E. Selenoprotein P: An extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu. Rev. Nutr. 2005, 25, 215–235.
- Qi, Y.; Grishin, N.V. Structural classification of thioredoxin-like fold proteins. Proteins 2005, 58, 376–388.
- Chivers, P.T.; Prehoda, K.E.; Raines, R.T. The CXXC motif: A rheostat in the active site. Biochemistry 1997, 36, 4061–4066.
- Chivers, P.T.; Laboissiere, M.C.; Raines, R.T. The CXXC motif: Imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996, 15, 2659–2667.
- Reeves, M.A.; Hoffmann, P.R. The human selenoproteome: Recent insights into functions and regulation. Cell. Mol. Life Sci. 2009, 66, 2457–2478.
- Lee, B.C.; Peterfi, Z.; Hoffmann, F.W.; Moore, R.E.; Kaya, A.; Avanesov, A.; Tarrago, L.; Zhou, Y.; Weerapana, E.; Fomenko, D.E.; et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol. Cell. 2013, 51, 397–404.
- Hawkes, W.C.; Alkan, Z. Regulation of redox signaling by selenoproteins. Biol. Trace Elem. Res. 2010, 134, 235–251.
- Martinez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Mas, D.; Valdivie, M.; Hu, C.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino. Acids. 2017, 49, 2091–2098.
- Bin, P.; Huang, R.; Zhou, X. Oxidation Resistance of the Sulfur Amino Acids: Methionine and Cysteine. Biomed. Res. Int. 2017, 2017, 9584932.
- Jiang, B.; Moskovitz, J. The Functions of the Mammalian Methionine Sulfoxide Reductase System and Related Diseases. Antioxidants 2018, 7, 122.
- Hansel, A.; Heinemann, S.H.; Hoshi, T. Heterogeneity and function of mammalian MSRs: Enzymes for repair, protection and regulation. J. Nutr. Biochem. 2005, 1703, 239–247.
- Cao, L.; Zhang, L.; Zeng, H.; Wu, R.T.; Wu, T.L.; Cheng, W.H. Analyses of Selenotranscriptomes and Selenium Concentrations in Response to Dietary Selenium Deficiency and Age Reveal Common and Distinct Patterns by Tissue and Sex in Telomere-Dysfunctional Mice. J. Nutr. 2017, 147, 1858–1866.
- Novoselov, S.V.; Kim, H.-Y.; Hua, D.; Lee, B.C.; Astle, C.M.; Harrison, D.E.; Friguet, B.; Moustafa, M.E.; Carlson, B.A.; Hatfield, D.L. Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid. Redox Signal. 2010, 12, 829–838.
- Gladyshev, V.N.; Stadtman, T.C.; Hatfield, D.L.; Jeang, K.T. Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc. Natl. Acad. Sci. USA 1999, 96, 835–839.
- Lourenço dos Santos, S.; Petropoulos, I.; Friguet, B. The Oxidized Protein Repair Enzymes Methionine Sulfoxide Reductases and Their Roles in Protecting against Oxidative Stress, in Ageing and in Regulating Protein Function. Antioxidants 2018, 7, 191.
- Kaya, A.; Lee, B.C.; Gladyshev, V.N. Regulation of protein function by reversible methionine oxidation and the role of selenoprotein MsrB1. Antioxid. Redox Signal. 2015, 23, 814–822.
- Hung, R.J.; Spaeth, C.S.; Yesilyurt, H.G.; Terman, J.R. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat. Cell. Biol. 2013, 15, 1445–1454.
- Kawabata Galbraith, K.; Kengaku, M. Multiple roles of the actin and microtubule-regulating formins in the developing brain. Neurosci. Res 2019, 138, 59–69.
- Tang, D.D. The Dynamic Actin Cytoskeleton in Smooth Muscle. Adv. Pharmacol 2018, 81, 1–38.
- Gallop, J.L. Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell. Dev. Biol. 2019.
- Leinweber, B.D.; Leavis, P.C.; Grabarek, Z.; Wang, C.-L.A.; Morgan, K.G. Extracellular regulated kinase (ERK) interaction with actin and the calponin homology (CH) domain of actin-binding proteins. Biochem. J. 1999, 344, 117–123.
- Fomenko, D.E.; Novoselov, S.V.; Natarajan, S.K.; Lee, B.C.; Koc, A.; Carlson, B.A.; Lee, T.H.; Kim, H.Y.; Hatfield, D.L.; Gladyshev, V.N. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: Roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J. Biol. Chem. 2009, 284, 5986–5993.
- Kim, K.Y.; Kwak, G.H.; Singh, M.P.; Gladyshev, V.N.; Kim, H.Y. Selenoprotein MsrB1 deficiency exacerbates acetaminophen-induced hepatotoxicity via increased oxidative damage. Arch. Biochem. Biophys. 2017, 634, 69–75.
- Jia, Y.; Zhou, J.; Liu, H.; Huang, K. Effect of methionine sulfoxide reductase B1 (SelR) gene silencing on peroxynitrite-induced F-actin disruption in human lens epithelial cells. Biochem. Biophys. Res. Commun. 2014, 443, 876–881.
- Dai, J.; Liu, H.; Zhou, J.; Huang, K. Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2016, 17, 231.
- Lee, B.C.; Lee, S.G.; Choo, M.K.; Kim, J.H.; Lee, H.M.; Kim, S.; Fomenko, D.E.; Kim, H.Y.; Park, J.M.; Gladyshev, V.N. Selenoprotein MsrB1 promotes anti-inflammatory cytokine gene expression in macrophages and controls immune response in vivo. Sci. Rep. 2017, 7, 5119.
- Achilli, C.; Ciana, A.; Minetti, G. Brain, immune system and selenium: A starting point for a new diagnostic marker for Alzheimer’s disease? Oxid. Med. Cell. Longev. 2018, 138, 223–226.
- He, Q.; Li, H.; Meng, F.; Sun, X.; Feng, X.; Chen, J.; Li, L.; Liu, J. Methionine Sulfoxide Reductase B1 Regulates Hepatocellular Carcinoma Cell Proliferation and Invasion via the Mitogen-Activated Protein Kinase Pathway and Epithelial-Mesenchymal Transition. Oxid. Med. Cell. Longev. 2018, 2018, 5287971.
- Li, H.; He, Q.; Meng, F.; Feng, X.; Chen, J.; Li, L.; Liu, J. Methionine sulfoxide reductase B1 regulates proliferation and invasion by affecting mitogen-activated protein kinase pathway and epithelial-mesenchymal transition in u2os cells. Biochem. Biophys. Res. Commun. 2018, 496, 806–813.
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443.
- Han, S.J.; Lee, B.C.; Yim, S.H.; Gladyshev, V.N.; Lee, S.R. Characterization of mammalian selenoprotein o: A redox-active mitochondrial protein. PLoS One 2014, 9, e95518.
- Sreelatha, A.; Yee, S.S.; Lopez, V.A.; Park, B.C.; Kinch, L.N.; Pilch, S.; Servage, K.A.; Zhang, J.; Jiou, J.; Karasiewicz-Urbanska, M.; et al. Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell 2018, 175, 809–821.e819.
- Dudkiewicz, M.; Szczepinska, T.; Grynberg, M.; Pawlowski, K. A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS One 2012, 7, e32138.
- Yan, J.; Fei, Y.; Han, Y.; Lu, S. Selenoprotein O deficiencies suppress chondrogenic differentiation of ATDC5 cells. Cell Biol. Int. 2016.




