Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Junhu Tai | + 3337 word(s) | 3337 | 2021-05-24 10:46:30 | | | |
| 2 | Peter Tang | Meta information modification | 3337 | 2021-06-02 05:08:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tai, J. Microbiota and Nasal Mucosal Diseases. Encyclopedia. Available online: https://encyclopedia.pub/entry/10395 (accessed on 08 February 2026).
Tai J. Microbiota and Nasal Mucosal Diseases. Encyclopedia. Available at: https://encyclopedia.pub/entry/10395. Accessed February 08, 2026.
Tai, Junhu. "Microbiota and Nasal Mucosal Diseases" Encyclopedia, https://encyclopedia.pub/entry/10395 (accessed February 08, 2026).
Tai, J. (2021, June 02). Microbiota and Nasal Mucosal Diseases. In Encyclopedia. https://encyclopedia.pub/entry/10395
Tai, Junhu. "Microbiota and Nasal Mucosal Diseases." Encyclopedia. Web. 02 June, 2021.
Copy Citation
The pathogenesis of nasal inflammatory diseases is related to various factors such as anatomical structure, heredity, and environment. The nasal microbiota play a key role in coordinating immune system functions. Dysfunction of the microbiota has a significant impact on the occurrence and development of nasal inflammation.
microbiota
nasal mucosa
immunity
chronic rhinosinusitis
allergic rhinitis
1. Introduction
In 2020, a panel of international experts, with more than 100 representatives from all over the world, defined the term “microbiota” [1]. They concluded that microbiota are composed of prokaryotes and eukaryotes, and that they are active within the range of microbial structures, metabolites, and movable genetic elements. Whipps et al. [2] first defined the term “microbiome” in 1988. Microbiomes encompass a wider range than microbiota. Phages, viruses, plasmids, free DNA, and a number of others such as prions and viroids are not considered to be members of the microbiota, but are included in the microbiome [3].
Many studies were conducted regarding gut microbiota, involving the most intensive and diverse microbial communities, which represent approximately 1.5 kg of bacteria in the gut [4][5]. pH of the gut, bile acids, and components of the innate immune system operate together to select and identify members of the gut microbiota [6][7][8]. Gut microbiota play a significant role in four domains of the human body. First, with regard to metabolism, gut microbiota can use dietary fiber and undigested proteins to provide energy and a variety of nutrients for the human body. Second, gut microbiota can protect the gut by secreting antimicrobial peptides, secretory IgA, and short-chain fatty acids. Third, they can also upregulate the expression of tight junction proteins, thereby improving the gut structure. Fourth, the gut is connected to the brain through the enteric nervous system (ENS), and neurotransmitters produced by gut microbiota participate in a variety of nervous systems [9]. Recently, various studies reported that changes in the gut microbiota are related to multiple disorders such as inflammatory bowel disease [10], Crohn’s disease [11], hepatitis C [12], Alzheimer’s disease [13], and depression [14].
Although the relevant mechanisms are still being studied, increasing evidence emphasizes the effect of gut microbiota on lung immunity, which is referred to as the gut-lung axis [15]. Bacterial community in the lungs is similar with that in the mouth. Streptococcus, Prevotella, and Veronica are the most commonly encountered genera [16]. Components of the pulmonary microbiota are thought to be transported from the oropharynx through micro-inhalation events and by mucosal diffusion involving adjacent tissues [17]. Various studies showed that changes in microbiota have a certain impact on the immunity of the lower airway mucosa. Eosinophilic inflammation, TH17 gene expression, neutrophilic inflammation, and the markers of allergic inflammation are all related to the differences in airway microbiota composition [18]. A report concluded that Staphylococcus, Propionibacterium, Corynebacterium, and Streptococcus are common bacterial genera in the nasal mucosal diseases of chronic rhinosinusitis (CRS) and allergic rhinitis (AR) [19]. In the upper respiratory tract immune system, microbiota are tolerated due to the low reactivity of the host immune system [20], and dysbiosis in the microbiome results in diseases of the upper respiratory tract, similarly to that in other human body sites [21].
Human nasal mucosa is the first contact point of inhaled environmental insults. Just as gut microbiota can protect the intestinal mucosa through immune regulation, microbiota in nasal mucosa are likely to play an important role in mucosal immunity. Although much research was conducted concerning the role of microbiota in lower respiratory tract disorders such as asthma, the role of microbiota in the upper respiratory tract including human nasal mucosa has not been studied in detail, especially with respect to immunity.
2. CRS and Microbiota
2.1. CRS Classification
CRS is a chronic inflammatory disease that occurs in the nasal cavity and sinuses and affects 12% of the global population [22]. The phenotypic classification of CRS is mainly based on the presence or absence of nasal polyps, which can be divided into CRS with nasal polyps (CRSwNP) or CRS without nasal polyps (CRSsNP) [23]. In contrast to the phenotype, the endotypic classification of CRS mainly represents an individual’s inflammatory mechanisms, rather than a clear entity with a direct biological basis. In a very meaningful study, researchers used 14 different inflammatory markers for hierarchical cluster analysis to determine the putative inflammatory endotype of CRS, and identified ten different clusters, including eosinophils and T helper-(Th) 2 related markers such as interleukin (IL)-5 and immunoglobulin E (IgE), neutrophils, or proinflammatory mediators such as IL-1β, IL-6, IL-8, and myeloperoxidase; Th17/Th22 markers such as IL-17A, IL-22, and tumor necrosis factor-α (TNF-α); and interferon-γ (IFN-γ) [24]. In recent years, a deeper understanding of the role of microbiota in the human immune system evolved; various inflammatory diseases, such as CRS, were reported to be associated with a significant shift in host microbiota from a healthy state to a diseased state [25]. Compared with that on other disorders such as asthma, research concerning microbiota and nasal diseases is still in its infancy, and causal relationships involving the existence of microbial communities and the development of CRS cannot be readily explained [26].
There are different types of microbiota in the upper airway of healthy adults (Figure 1). The nasal cavity is directly connected to the external environment. Through inhalation, the nasal cavity can directly contact various microbiota, fungal spores, and pollutants [27]. The microbiota of healthy adults’ anterior nares is mainly composed of Actinobacteria, Firmicutes, and Proteobactera [28]. Researchers examined the anterior nares of 236 healthy adults using nasal swabs and concluded that Staphylococcus, Propionibacterium, Corynebacterium, and Moraxella were the most common microbiota in their anterior nares [29]. One study concluded that tissue samples were more suitable for assessing microbiological groups in CRS patients than nasal swabs, because they observed significant differences in the microbiota groups in the nasal swabs, while the differences observed in the tissue samples were smaller [30]. However, there are some discrepancies about the usefulness of the two methods. [31]. Other data showed that a tissue biopsy cannot provide additional information compared with multiple swab tests. In more than 90% of their cases, swabs from multiple sites provide comprehensive information about patients’ culturable pathogens. In the middle meatus of healthy adults, the most abundant microbiota were Staphylococcus aureus (S. aureus), Staphylococcus epidermidis, and Propionibacterium acnes [32]. Using next-generation 454 pyrosequencing of the 16S rRNA gene, Jetté et al. found that Streptococcus, Prevotella, Veillonella, and Haemophilus were the most common microbiota in the throats of 97 adults [33]. The common microbiota in CRS patients vary with geographical location. According to data reported, Cyanobacteria are the dominant phylum in CRS in Missouri, USA, and that the change in microbiota composition between the control group and CRS group is minor [34]; in contrast, other data show that the abundance of Verrucomicrobia and Bacteroides is low and that of Actinobacteria is high in Colorado, USA [35]. A Korean study compared a CRS group with a control group and found that the abundance of Bacteroides in the CRS group was low and that of Fusobacteria was high [36].
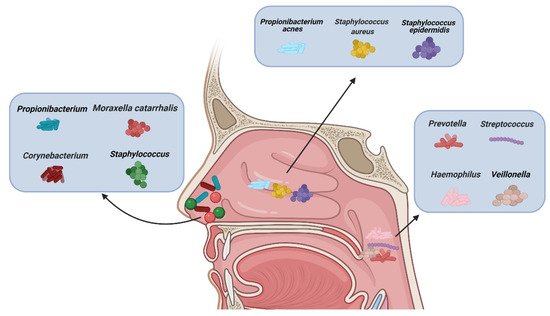
Figure 1. Types of microbiota in normal nasal mucosa and other parts of the upper airway. (Figure created with Biorender.com).
2.2. Type 2 CRS and Microbiota
Relationships between CRS and various microbiota cultured in the nasal system have been studied for many years [37]. Microbiotal dysbiosis is considered an important biomarker of CRS [38][39]. Recent advances in new detection methods aroused interest in the role of microbiota in long-term diseases, which can be used to identify previously unrecognizable, unculturable microbiota. Quantitative polymerase chain reaction (qPCR), fluorescence in situ hybridization (FISH), mass spectrometry, and DNA microarrays were used to identify microbiota and to visualize biofilms in clinical samples of patients with CRS [40]. The development of next-generation sequencing (NGS) provides a non-targeted molecular method. Specifically, 16S amplicon DNA sequencing is a NGS technique in which universal primers for the 16S rRNA gene are used; this suggests that bacterial organisms participate in the pathogenesis of CRS, and may indicate that disordering of normal microbiota community structures in the nasal sinus mucosa is one of the causes of CRS [41][42]. Metabolic exchange plays an important role in maintaining the interdependence between microbiota [43]. An outstanding study pointed out that Corynebacterium, one of the common nasal bacteria, inhibits the growth of Streptococcus pneumoniae by releasing triacylglycerol on the skin surface of the host [44]. Some studies also found that in CRS, the growth of S. aureus is often closely related to Staphylococcus epidermidis and Propionibacterium acnes [45]. A most recent study in South Korea obtained interesting data [46]. They found that the use of antibiotics can cause differences in secretory proteome according to the condition of the disease. Their data suggest that the use of antibiotics should be considered as a confounding factor in proteomics research.
There are various microbiota, such as Staphylococcus, Streptococcus, Propionibacterium, and Corynebacterium, (Figure 2A) in normal nasal mucosa [47]. However, the type and quantity of microbiota change significantly in the mucosa of patients with CRS (Figure 2B,C). Changes in microbiota are related to various factors. In addition to significant differences between subjects, age and smoking influence the composition and distribution of microbiotal species [48][49]. The frequent use of antibiotics may also induce instability in the microbiota. A previous study compared the microbiota of paranasal sinuses of patients with CRS before and after drug treatment and found that the diversity and uniformity of bacteria decreased significantly after high-dose antibiotic treatments [50]. In a cross-sectional study, surgery was shown to affect the microbiotal ecology of the sinuses, leading to a reduction in microbiota abundance [35]. Sinus surgery has a similar effect on fungal populations. The abundance and diversity of fungi in the sinus cavity of patients after endoscopic sinus surgery are significantly decreased [51][52]. The results of a meta-analysis [53] revealed that bacterial richness and diversity in CRS decreased, which supports the keystone-pathogen hypothesis; that is, certain pathogenic microbiota that usually exist in low abundance may form a microbiome under disease conditions [54]. Disruption of the microbial community leads to the loss of key symbiotic species. Under normal circumstances, these symbiotic species may prevent the excessive growth of pathogens, and the loss of variety and diversity of CRS microbiota seems to be the products of tissue eosinophilia and mucosal inflammation; whether this disorder is a cause or a consequence of an impaired epithelial integrity disease remains a subject for further research [55].
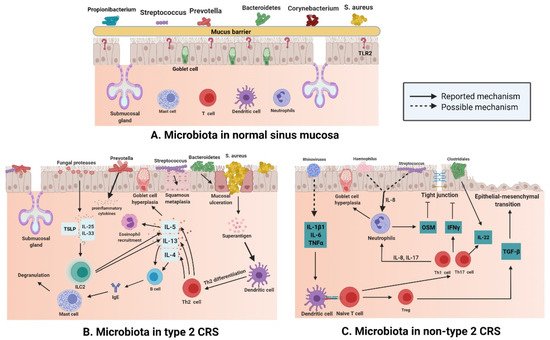
Figure 2. (A) Different kinds of microbiota colonize healthy nasal mucosa such as Staphylococcus, Streptococcus, Propionibacterium, and Corynebacterium. (B) With the loss of epithelial integrity, pattern recognition molecules decrease, which provides an environment for bacteria to enter through the mucosal barrier in type 2 CRS. Mucosal ulceration is associated with increased abundance of Bacteroides, while squamous metaplasia is associated with increased Streptococcus abundance; enterotoxins produced by S. aureus can act as superantigens and promote Th-2 inflammation, thereby leading to the production of cytokines, such as IL-13, IL-4, and IL-5, in Type 2 CRS. At the same time, Prevotella is related to the release of proinflammatory cytokines, and fungal proteases can induce the production of TSLP, which leads to the activation of ILC2s producing IL-5 and IL-13. (C) In non-type 2 CRS, an increase in the abundance of Haemophilus or Streptococcus may be related to elevated IL-8 levels and neutrophil counts. Rhinovirus may increase the levels of IL-1B1, IL-6, and TNF-α; induce dendritic cell differentiation; and boost IL-22. At the same time, increased abundance of Clostridiales also elevates IL-22 levels. (Figure created with Biorender.com).
S. aureus is a common type of microbiota in the nasal mucosa [56], but many studies have shown that compared with that in the normal nasal mucosa, the number of S. aureus in the patients with type 2 CRS is greatly increased (Figure 2B) [57]. S. aureus produces enterotoxins, and it can be recognized as superantigens and by Th-2 inflammation that is promoted by it; S. aureus also leads to the secretion of cytokines such as IL-13, IL-4, and IL-5 in type 2 CRS [58]. Mucosal ulceration is associated with an increased abundance of Bacteroides [59], squamous metaplasia associated with high Streptococcus levels [60]; Prevotella is related to the release of proinflammatory cytokines [61], and the production of thymic stromal lymphopoietin(TSLP) is related to the induction of fungal protease, which leads to the activation of type 2 innate lymphoid cells (ILC2s) producing IL-5 and IL-13 [62].
2.3. Non-Type 2 CRS and Microbiota
Non-type 2 CRS is a heterogeneous disease; additional definitions are needed to guide its correct diagnosis and treatment, as most CRS studies focus on type 2 CRS, and there is not enough information regarding the internal classification of non-type 2 CRS [63]. Diseases that can easily induce non-type 2 CRS include acute rhinosinusitis, which is usually caused by a viral respiratory tract infection, asthma, tonsillitis, bronchitis, allergic and non-allergic rhinitis, pneumonia, and gastroesophageal reflux disease [64]; however, potential susceptibility conditions, such as primary and secondary immunodeficiency, including HIV infection, cystic fibrosis, and cilia dyskinesia, should also be considered [65].
The number of eosinophils and plasma cells in the mucosa of non-type 2 CRS is less than that in type 2 CRS, but the number of neutrophils in the mucosa of non-type 2 CRS is higher than that in type 2 CRS (Figure 2C). Neutrophil inflammation of nasal mucosa is a characteristic of non-type 2 CRS, which is caused by infection or external stimuli; type 1 inflammation, which is based on the Th1 cell, and type 3 inflammation, which is based on the Th17 cell, are present in an equal proportion in non-type 2 CRS [66]. Invasion by external pathogens induces the secretion of IL-6, IL-8, and TNF-α in the nasal epithelium (Figure 2C), which could be caused by a rhinovirus [67] that activates dendritic cells (DCs). IL-8 secreted by epithelial cells recruits neutrophils, which cause goblet cells to proliferate and destroy tight junctions [68]. In 28 patients with asthma, terminal restriction fragment length polymorphism (T-RFLP) analysis showed that Moraxella, Haemophilus, and Streptococcus were the dominant species in the respiratory tract bacterial community, and the total abundance of these microbiota was significantly and positively correlated with the concentration of IL-8 and neutrophil count in sputum [69]. Lal et al. [47] conducted an inter-subject microbiotal analysis of 65 subjects. They found that the diversity of microbiota in patients with non-type 2 CRS was lower than that in the control group (healthy and AR subjects) or in patients with type 2 CRS. Fusobacterium, Propionibacterium, Haemophilus, and Streptococcus were the main bacteria in non-type 2 CRS patients. Therefore, it can be speculated that Haemophilus and Streptococcus may be involved in the secretion of IL-8 and recruitment of neutrophils in non-type 2 CRS (Figure 2). The reported data show that levels of IL-22 receptors are increased in non-type 2 CRS [70], and studies showed that IL-22 production in the gut is induced by Clostridium [71]. A combination of data from 51 patients with CRS suggested that the increase in IL-22 receptor levels in non-type 2 CRS may be a result of the predominance of Clostridium in nasal microbiota and IL-22 cytokine production [60].
3. AR and Microbiota
3.1. AR and Type 1 Hypersensitivity
AR is a common symptom of type 1 hypersensitivity and Th2-mediated inflammatory disease [72]. Epidemiological studies show that nearly a quarter of adults and almost half of children are affected [73]. AR was previously considered to be a disease confined to the nasal cavity; however, it is now considered to be a manifestation of systemic airway disease, which is usually comorbid with asthma [74]. As an IgE mediated type 1 hypersensitivity process, AR symptoms are caused by allergens, and when the nasal mucosa is directly or indirectly exposed to allergens such as mold, pollen, dust, and mite feces, innate immune cells and adaptive immune cells participate in the pathophysiological mechanisms involved in AR, inducing IgE production, eosinophil activation, mast cell recruitment, and basophils degranulation, and then present a variety of clinical symptoms of AR [75]. Therefore, minimizing allergen exposure should be an important part of any treatment plan [76]. In addition to avoiding known allergens, intranasal corticosteroids, which are one of the most effective therapeutics, should be used as the first-line treatment; however, when there is no response to intranasal corticosteroids, second-line treatment should be considered, including antihistamines, decongestants, leukotriene receptor antagonists, and non-drug treatments, such as nasal irrigation. Subcutaneous or sublingual immunotherapy should be considered if a patient’s AR symptoms cannot be fully controlled by conventional treatment modalities [77].
As time goes by, more and more achievements were made in elucidating the occurrence and mechanisms underlying anaphylaxis. The key processes involved in anaphylaxis include the activation and maturation of DCs after exposure to allergens; subsequently, the initial signals provided by mucosal epithelial cells and DCs lead to the cloning and expansion of allergen-specific Th2 cells, which are important driving factors in AR pathology [78]; it was shown that Th2 cells are related to the sensitization and staging of AR [79]. ILC2s are also activated by cytokines such as IL-25, IL-33, and TSLP. Th2 cells and ILC2s produce type 2 cells, including IL-4, IL-5, IL-13, IL-25, IL-33, and TSLP. IL-4 and IL-13 drive B cells to produce allergen-specific IgE, which can combine with mast cells. IL-5 contributes to eosinophil recruitment of eosinophils [80]. Some studies reported that serum IL-17 level is correlated with allergic severity during high pollen-level seasons, which is considered to be a marker of the severity of the allergy in patients with AR. Additionally, myeloid DCs isolated from patients with pollen allergy increase the tendency of inducing T cells to secrete IL-17 in vitro [81]. Viral infection may lead to the occurrence and aggravation of AR. During the common cold, mast cells congregate, leading to deterioration in allergic conditions; the key factors influencing such allergic reactions are stage, genetic background, gender, and age at viral infection [82].
3.2. AR and Microbiota
The incidence rate of allergic diseases is closely related to interactions between the host system and resident microbiota [83]. Some studies showed that symbiotic microbiota regulate susceptibility to allergic diseases, and the absence of symbiotic bacteria can enhance the proliferation of basophils, increase the number of infiltrating lymphocytes and eosinophils, aggravate Th2 cell reactions and allergic inflammation, and reduce the number of regulatory T (Treg) and Th17 cells [84][85].
This situation is similar to that encountered in normal sinus mucosa; S. aureus, Propionibacterium, Prevotella, Corynebacterium, Bacteroidetes, and Streptococcus are common in normal nasal mucosa (Figure 3A); however, the abundance of S. aureus, Propionibacterium, Corynebacterium, and peptoniphilus in the nasal mucosa of patients with AR is considerably increased compared with that of the common bacteria in nasal mucosa of normal individuals (Figure 3B), while the number of Prevotella and Streptococcus is decreased [47]. In a study of 20 patients with AR and 12 normal controls, the researchers used 454 pyrosequencing based on the 16S rRNA gene to describe and compare the inferior turbinate mucosal microbiota of normal controls and patients with AR, and found that the inferior turbinate microbiota imbalance in patients with AR was related to the total IgE level; their results emphasized the relationship between inferior turbinate microbiota imbalance and the onset of AR [86].
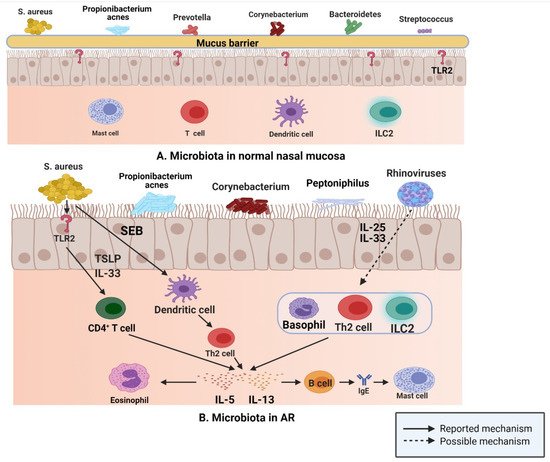
Figure 3. (A) S. aureus, Propionibacterium, Prevotella, Corynebacterium, Bacteroidetes, and Streptococcus are common in normal nasal mucosa. (B) In patients with AR, S. aureus can produce IL-5 and IL-13 by binding to TLR2, and SEB can induce the release of IL-5 and IL-13 by affecting Th2 cells. Rhinovirus stimulates human respiratory epithelial cells to produce IL-25 and IL-33, which drive the production of IL-5 and IL-13 by binding to Th2 cells, ILC2s, and basophils (Figure created with Biorender.com).
In the upper respiratory tract, microbiota play an important role in driving type 2 immune responses, according to the data of a study published in 2016; after binding with toll like receptor (TLR) 2, S. aureus induces the production of type 2 cytokines, such as IL-5 and IL-13, via IL-33 released from human airway epithelial cells and TSLP [87]. Furthermore, staphylococcal enterotoxin B (SEB) induces IL-5 and IL13 release by affecting Th2 cells [88]. Rhinovirus, one of the most common viruses in the human respiratory tract, is closely related to the occurrence and development of allergic asthma and plays a key role in the propagation of the type 2 immune response. IL-25 and IL-33 are produced by human respiratory epithelial cells stimulated by rhinovirus, which then drive the production of IL-5 and IL-13 by binding to the receptors on Th2 cells, ILC2s, and basophils [89][90]. These released type 2 cytokines are actively involved in the type 2 immune response. IL-5 participates in the recruitment of eosinophils and is related to their development and activation [91]. IL-13 upregulates class II expression in B cells and promotes IgE class conversion, and then IgE binds to mast cell receptors [92].
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103.
- Whipps, J.L.K.; Cooke, R. Mycoparasitism and plant disease control. In Fungi in Biological Control Systems; Burge, N.M., Ed.; Manchester University Press: Manchester, UK, 1988; pp. 161–187.
- Dupré, J.O.M.; Maureen, A. Varieties of living things life at the intersection of lineage and metabolism. In Vitalism and the Scientific Image in Post-Enlightenment Life Science; History, Philosophy and Theory of the Life Sciences; Normandin, S., Wolfe, C., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 1800–2010.
- Carvalho, B.M.; Guadagnini, D.; Tsukumo, D.M.L.; Schenka, A.A.; Latuf-Filho, P.; Vassallo, J.; Dias, J.C.; Kubota, L.T.; Carvalheira, J.B.C.; Saad, M.J.A. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 2012, 55, 2823–2834.
- Nicholson, J.K.; Holmes, E.; Wilson, I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005, 3, 431–438.
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108.
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235.
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273.
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. CMLS 2019, 76, 473–493.
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10.
- Torres, J.; Mehandru, S.; Colombel, J.F.; Peyrin-Biroulet, L. Crohn’s disease. Lancet 2017, 389, 1741–1755.
- Preveden, T.; Scarpellini, E.; Milić, N.; Luzza, F.; Abenavoli, L. Gut microbiota changes and chronic hepatitis C virus infection. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 813–819.
- Jiang, C.; Li, G.; Huang, P.; Liu, Z.; Zhao, B. The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers Dis. JAD 2017, 58, 1–15.
- Koopman, M.; El Aidy, S. Depressed gut? The microbiota-diet-inflammation trialogue in depression. Curr. Opin. Psychiatry 2017, 30, 369–377.
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The Gut-Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. 2), S150–S156.
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075.
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. Homeostasis and its disruption in the lung microbiome. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L1047–L1055.
- Ver Heul, A.; Planer, J.; Kau, A.L. The Human Microbiota and Asthma. Clin. Rev. Allergy Immunol. 2019, 57, 350–363.
- Lee, J.T.; Kim, C.M.; Ramakrishnan, V. Microbiome and disease in the upper airway. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 1–6.
- Shroff, K.E.; Meslin, K.; Cebra, J.J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 1995, 63, 3904–3913.
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87.
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 58, 1–464.
- Bauer, A.M.; Turner, J.H. Personalized Medicine in Chronic Rhinosinusitis: Phenotypes, Endotypes, and Biomarkers. Immunol. Allergy Clin. N. Am. 2020, 40, 281–293.
- Tomassen, P.; Vandeplas, G.; Van Zele, T.; Cardell, L.O.; Arebro, J.; Olze, H.; Förster-Ruhrmann, U.; Kowalski, M.L.; Olszewska-Ziąber, A.; Holtappels, G.; et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J. Allergy Clin. Immunol. 2016, 137, 1449–1456.e4.
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141.
- Ramakrishnan, V.R.; Frank, D.N. Microbiome in patients with upper airway disease: Moving from taxonomic findings to mechanisms and causality. J. Allergy Clin. Immunol. 2018, 142, 73–75.
- Rawls, M.; Ellis, A.K. The microbiome of the nose. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2019, 122, 17–24.
- Bassis, C.M.; Tang, A.L.; Young, V.B.; Pynnonen, M.A. The nasal cavity microbiota of healthy adults. Microbiome 2014, 2, 27.
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E.; et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 15, R66.
- Cho, S.W.; Kim, D.Y.; Choi, S.; Won, S.; Kang, H.R.; Yi, H. Microbiome profiling of uncinate tissue and nasal polyps in patients with chronic rhinosinusitis using swab and tissue biopsy. PLoS ONE 2021, 16, e0249688.
- Szaleniec, J.; Gibała, A.; Hartwich, P.; Hydzik-Sobocińska, K.; Konior, M.; Gosiewski, T.; Szaleniec, M. Challenging the gold standard: Methods of sampling for microbial culture in patients with chronic rhinosinusitis. Eur. Arch. Oto-Rhino-Laryngol. Off. J. Eur. Fed. Oto-Rhino-Laryngol. Soc. (EUFOS) Affil. Ger. Soc. Oto-Rhino-Laryngol. Head Neck Surg. 2021.
- Ramakrishnan, V.R.; Feazel, L.M.; Gitomer, S.A.; Ir, D.; Robertson, C.E.; Frank, D.N. The microbiome of the middle meatus in healthy adults. PLoS ONE 2013, 8, e85507.
- Jetté, M.E.; Dill-McFarland, K.A.; Hanshew, A.S.; Suen, G.; Thibeault, S.L. The human laryngeal microbiome: Effects of cigarette smoke and reflux. Sci. Rep. 2016, 6, 35882.
- Aurora, R.; Chatterjee, D.; Hentzleman, J.; Prasad, G.; Sindwani, R.; Sanford, T. Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol. Head Neck Surg. 2013, 139, 1328–1338.
- Feazel, L.M.; Robertson, C.E.; Ramakrishnan, V.R.; Frank, D.N. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope 2012, 122, 467–472.
- Choi, E.B.; Hong, S.W.; Kim, D.K.; Jeon, S.G.; Kim, K.R.; Cho, S.H.; Gho, Y.S.; Jee, Y.K.; Kim, Y.K. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy 2014, 69, 517–526.
- Mahdavinia, M.; Keshavarzian, A.; Tobin, M.C.; Landay, A.L.; Schleimer, R.P. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2016, 46, 21–41.
- Copeland, E.; Leonard, K.; Carney, R.; Kong, J.; Forer, M.; Naidoo, Y.; Oliver, B.G.G.; Seymour, J.R.; Woodcock, S.; Burke, C.M.; et al. Chronic Rhinosinusitis: Potential Role of Microbial Dysbiosis and Recommendations for Sampling Sites. Front. Cell. Infect. Microbiol. 2018, 8, 57.
- Hoggard, M.; Biswas, K.; Zoing, M.; Wagner Mackenzie, B.; Taylor, M.W.; Douglas, R.G. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2017, 7, 230–239.
- Jervis Bardy, J.; Psaltis, A.J. Next Generation Sequencing and the Microbiome of Chronic Rhinosinusitis: A Primer for Clinicians and Review of Current Research, Its Limitations, and Future Directions. Ann. Otol. Rhinol. Laryngol. 2016, 125, 613–621.
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764.
- Lam, K.; Schleimer, R.; Kern, R.C. The Etiology and Pathogenesis of Chronic Rhinosinusitis: A Review of Current Hypotheses. Curr. Allergy Asthma Rep. 2015, 15, 41.
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454.
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. mBio 2016, 7, e01725-15.
- Kaspar, U.; Kriegeskorte, A.; Schubert, T.; Peters, G.; Rudack, C.; Pieper, D.H.; Wos-Oxley, M.; Becker, K. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 2016, 18, 2130–2142.
- Kim, Y.S.; Han, D.; Mo, J.H.; Kim, Y.M.; Kim, D.W.; Choi, H.G.; Park, J.W.; Shin, H.W. Antibiotic-Dependent Relationships Between the Nasal Microbiome and Secreted Proteome in Nasal Polyps. Allergy Asthma Immunol. Res. 2021, 13, e46.
- Lal, D.; Keim, P.; Delisle, J.; Barker, B.; Rank, M.A.; Chia, N.; Schupp, J.M.; Gillece, J.D.; Cope, E.K. Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int. Forum Allergy Rhinol. 2017, 7, 561–569.
- Ramakrishnan, V.R.; Feazel, L.M.; Abrass, L.J.; Frank, D.N. Prevalence and abundance of Staphylococcus aureus in the middle meatus of patients with chronic rhinosinusitis, nasal polyps, and asthma. Int. Forum Allergy Rhinol. 2013, 3, 267–271.
- Yu, G.; Phillips, S.; Gail, M.H.; Goedert, J.J.; Humphrys, M.S.; Ravel, J.; Ren, Y.; Caporaso, N.E. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 2017, 5, 3.
- Liu, C.M.; Soldanova, K.; Nordstrom, L.; Dwan, M.G.; Moss, O.L.; Contente-Cuomo, T.L.; Keim, P.; Price, L.B.; Lane, A.P. Medical therapy reduces microbiota diversity and evenness in surgically recalcitrant chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2013, 3, 775–781.
- Murr, A.H.; Goldberg, A.N.; Pletcher, S.D.; Dillehay, K.; Wymer, L.J.; Vesper, S.J. Some chronic rhinosinusitis patients have elevated populations of fungi in their sinuses. Laryngoscope 2012, 122, 1438–1445.
- Cleland, E.J.; Bassiouni, A.; Boase, S.; Dowd, S.; Vreugde, S.; Wormald, P.J. The fungal microbiome in chronic rhinosinusitis: Richness, diversity, postoperative changes and patient outcomes. Int. Forum Allergy Rhinol. 2014, 4, 259–265.
- Wagner Mackenzie, B.; Waite, D.W.; Hoggard, M.; Douglas, R.G.; Taylor, M.W.; Biswas, K. Bacterial community collapse: A meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ. Microbiol. 2017, 19, 381–392.
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725.
- Rom, D.; Bassiouni, A.; Eykman, E.; Liu, Z.; Paramasivan, S.; Alvarado, R.; Earls, P.; Psaltis, A.J.; Harvey, R.J. The Association Between Disease Severity and Microbiome in Chronic Rhinosinusitis. Laryngoscope 2019, 129, 1265–1273.
- Yan, M.; Pamp, S.J.; Fukuyama, J.; Hwang, P.H.; Cho, D.Y.; Holmes, S.; Relman, D.A. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 2013, 14, 631–640.
- Boase, S.; Foreman, A.; Cleland, E.; Tan, L.; Melton-Kreft, R.; Pant, H.; Hu, F.Z.; Ehrlich, G.D.; Wormald, P.J. The microbiome of chronic rhinosinusitis: Culture, molecular diagnostics and biofilm detection. BMC Infect. Dis. 2013, 13, 210.
- Van Zele, T.; Gevaert, P.; Watelet, J.B.; Claeys, G.; Holtappels, G.; Claeys, C.; van Cauwenberge, P.; Bachert, C. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. J. Allergy Clin. Immunol. 2004, 114, 981–983.
- Ramakrishnan, V.R.; Hauser, L.J.; Feazel, L.M.; Ir, D.; Robertson, C.E.; Frank, D.N. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J. Allergy Clin. Immunol. 2015, 136, 334–342.e331.
- Kuhar, H.N.; Tajudeen, B.A.; Mahdavinia, M.; Heilingoetter, A.; Ganti, A.; Gattuso, P.; Ghai, R.; Batra, P.S. Relative abundance of nasal microbiota in chronic rhinosinusitis by structured histopathology. Int. Forum Allergy Rhinol. 2018, 8, 1430–1437.
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The Human Microbiome during Bacterial Vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238.
- Kouzaki, H.; O’Grady, S.M.; Lawrence, C.B.; Kita, H. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. J. Immunol. 2009, 183, 1427–1434.
- Payne, S.C.; Borish, L.; Steinke, J.W. Genetics and phenotyping in chronic sinusitis. J. Allergy Clin. Immunol. 2011, 128, 710–720; quiz 721–722.
- Lam, K.; Hirsch, A.G.; Tan, B.K. The association of premorbid diseases with chronic rhinosinusitis with and without polyps. Curr. Opin. Otolaryngol. Head Neck Surg. 2014, 22, 231–241.
- Cho, S.H.; Kim, D.W.; Gevaert, P. Chronic Rhinosinusitis without Nasal Polyps. J. Allergy Clin. Immunol. Pract. 2016, 4, 575–582.
- Lee, K.; Tai, J.; Lee, S.H.; Kim, T.H. Advances in the Knowledge of the Underlying Airway Remodeling Mechanisms in Chronic Rhinosinusitis Based on the Endotypes: A Review. Int. J. Mol. Sci. 2021, 22, 910.
- van Kempen, M.; Bachert, C.; Van Cauwenberge, P. An update on the pathophysiology of rhinovirus upper respiratory tract infections. Rhinology 1999, 37, 97–103.
- Bachert, C.; van Kempen, M.J.; Höpken, K.; Holtappels, G.; Wagenmann, M. Elevated levels of myeloperoxidase, pro-inflammatory cytokines and chemokines in naturally acquired upper respiratory tract infections. Eur. Arch. Oto-Rhino-Laryngol. Off. J. Eur. Fed. Oto-Rhino-Laryngol. Soc. (EUFOS) Affil. Ger. Soc. Oto-Rhino-Laryngol. Head Neck Surg. 2001, 258, 406–412.
- Green, B.J.; Wiriyachaiporn, S.; Grainge, C.; Rogers, G.B.; Kehagia, V.; Lau, L.; Carroll, M.P.; Bruce, K.D.; Howarth, P.H. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE 2014, 9, e100645.
- Detwiller, K.Y.; Smith, T.L.; Alt, J.A.; Trune, D.R.; Mace, J.C.; Sautter, N.B. Differential expression of innate immunity genes in chronic rhinosinusitis. Am. J. Rhinol. Allergy 2014, 28, 374–377.
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150.
- Scadding, G. Cytokine profiles in allergic rhinitis. Curr. Allergy Asthma Rep. 2014, 14, 435.
- Hoyte, F.C.L.; Nelson, H.S. Recent advances in allergic rhinitis. F1000Research 2018, 7.
- Morjaria, J.B.; Caruso, M.; Emma, R.; Russo, C.; Polosa, R. Treatment of Allergic Rhinitis as a Strategy for Preventing Asthma. Curr. Allergy Asthma Rep. 2018, 18, 23.
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454.
- Kakli, H.A.; Riley, T.D. Allergic Rhinitis. Prim. Care 2016, 43, 465–475.
- Sur, D.K.; Plesa, M.L. Treatment of Allergic Rhinitis. Am. Fam. Physician 2015, 92, 985–992.
- Meng, Y.; Wang, C.; Zhang, L. Recent developments and highlights in allergic rhinitis. Allergy 2019, 74, 2320–2328.
- Iinuma, T.; Okamoto, Y.; Morimoto, Y.; Arai, T.; Sakurai, T.; Yonekura, S.; Sakurai, D.; Hirahara, K.; Nakayama, T. Pathogenicity of memory Th2 cells is linked to stage of allergic rhinitis. Allergy 2018, 73, 479–489.
- van de Veen, W.; Akdis, M. The use of biologics for immune modulation in allergic disease. J. Clin. Investig. 2019, 129, 1452–1462.
- Ciprandi, G.; De Amici, M.; Murdaca, G.; Fenoglio, D.; Ricciardolo, F.; Marseglia, G.; Tosca, M. Serum interleukin-17 levels are related to clinical severity in allergic rhinitis. Allergy 2009, 64, 1375–1378.
- Tantilipikorn, P. The relationship between allergic rhinitis and viral infections. Curr. Opin. Otolaryngol. Head Neck Surg. 2014, 22, 249–252.
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323.
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt⁺ T cells. Science 2015, 349, 989–993.
- Hill, D.A.; Siracusa, M.C.; Abt, M.C.; Kim, B.S.; Kobuley, D.; Kubo, M.; Kambayashi, T.; Larosa, D.F.; Renner, E.D.; Orange, J.S.; et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012, 18, 538–546.
- Hyun, D.W.; Min, H.J.; Kim, M.S.; Whon, T.W.; Shin, N.R.; Kim, P.S.; Kim, H.S.; Lee, J.Y.; Kang, W.; Choi, A.M.K.; et al. Dysbiosis of Inferior Turbinate Microbiota Is Associated with High Total IgE Levels in Patients with Allergic Rhinitis. Infect. Immun. 2018, 86.
- Lan, F.; Zhang, N.; Gevaert, E.; Zhang, L.; Bachert, C. Viruses and bacteria in Th2-biased allergic airway disease. Allergy 2016, 71, 1381–1392.
- Gorski, S.A.; Hahn, Y.S.; Braciale, T.J. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog. 2013, 9, e1003615.
- Beale, J.; Jayaraman, A.; Jackson, D.J.; Macintyre, J.D.R.; Edwards, M.R.; Walton, R.P.; Zhu, J.; Man Ching, Y.; Shamji, B.; Edwards, M.; et al. Rhinovirus-induced IL-25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci. Transl. Med. 2014, 6, 256ra134.
- Shikotra, A.; Choy, D.F.; Ohri, C.M.; Doran, E.; Butler, C.; Hargadon, B.; Shelley, M.; Abbas, A.R.; Austin, C.D.; Jackman, J.; et al. Increased expression of immunoreactive thymic stromal lymphopoietin in patients with severe asthma. J. Allergy Clin. Immunol. 2012, 129, 104–111.
- Sitkauskiene, B.; Johansson, A.K.; Sergejeva, S.; Lundin, S.; Sjöstrand, M.; Lötvall, J. Regulation of bone marrow and airway CD34+ eosinophils by interleukin-5. Am. J. Respir. Cell Mol. Biol. 2004, 30, 367–378.
- Wynn, T.A. IL-13 effector functions. Annu. Rev. Immunol. 2003, 21, 425–456.
More
Information
Subjects:
Endocrinology & Metabolism
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
02 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




