
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pamela Acha | + 2762 word(s) | 2762 | 2021-05-24 10:40:24 | | | |
| 2 | Peter Tang | Meta information modification | 2762 | 2021-05-31 10:13:29 | | |
Video Upload Options
Myelodysplastic/myeloproliferative neoplasms (MDS/MPN) are myeloid neoplasms characterized by the presentation of overlapping features from both myelodysplastic syndromes and myeloproliferative neoplasms. Although the classification of MDS/MPN relies largely on clinical features and peripheral blood and bone marrow morphology, studies have demonstrated that a large proportion of patients (~90%) with this disease harbor somatic mutations in a group of genes that are common across myeloid neoplasms. These mutations play a role in the clinical heterogeneity of these diseases and their clinical evolution.
1. Introduction
2. Diagnostic Criteria of MDS/MPN
3. Cytogenetic Abnormalities in MDS/MPN
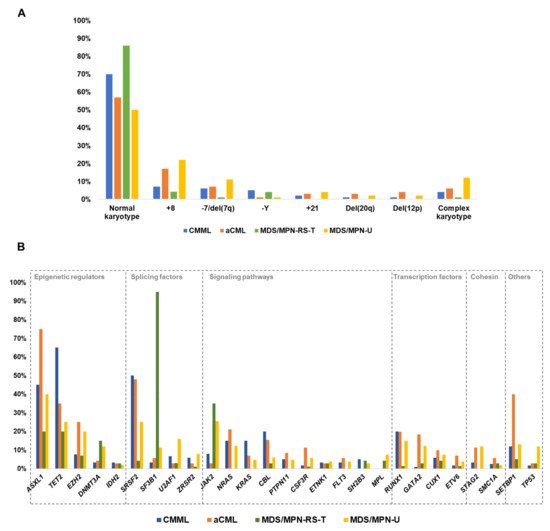
|
MDS/MPN |
Abnormal Karyotypes (%) |
Common Abnormalities (Frequency %) |
|---|---|---|
|
CMML |
30% |
+8: 6–7% −Y: 4–6% −7/del(7q): 2–9% +21: 1–2% CK: 3–6% Deletions of 20q (1–2%) and 12p (1%) |
|
aCML |
43% |
+8: 17% −7/del(7q): 6–8% CK: 4–8% |
|
MDS/MPN-RS-T |
10–17% |
+8: 4% −Y: 4% CK: 0–4% |
|
MDS/MPN-U |
50% |
+8: 14–25% −7/del(7q): 11% CK: 12% |
|
JMML |
19–35% |
−7: 9–25% Others (del(7q), +8): 10% |
|
MDS/MPN Subtype |
Diagnosis |
Prognosis |
|---|---|---|
|
CMML |
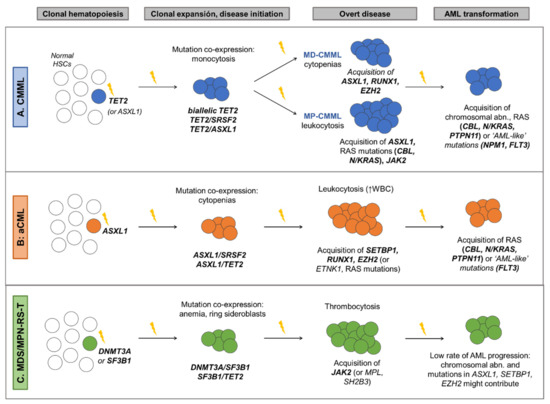
-WHO [1]: presence of mutations in genes often associated with CMML (TET2, SRSF2, ASXL1, SETBP1) in the proper clinical contest can be used to support diagnosis -Associated with the following gene mutation combinations: TET2-SRSF2, biallelic TET2, SRSF2-RUNX1 [2][4][30] |
Cytogenetics -Three cytogenetic stratification systems have been proposed [23][24][25] -Recurrent findings: • Low risk karyotypes: normal karyotype, isolated loss of Y • High risk karyotypes: chr7 abnormalities, complex karyotype, monosomal karyotype Gene mutations: -Unfavorable outcome: mutations in ASXL1, RUNX1, NRAS and SETBP1 [2][30][31] -Favorable outcome: TET2 mutations, especially in the absence of ASXL1 mutations (TET2MUT/ASXL1WT). These patients also show better response to HMA [32][33][34]. Prognostic stratification: -GFM Model [2], stratification in 3 risk groups based on: Age > 65 years; Hb < 10 g/dL in females and <11 g/dL in males; WBC > 15 × 109/L; Platelet count < 100 × 109/L; ASXL1 mutations -Mayo Molecular Model (MMM) [31], stratification in 4 risk groups based on: Hb < 10 g/dL; AMC > 10 × 109/L; Platelet count < 100 × 109/L; Presence of circulating IMCs; ASXL1 mutations -CPSS-Mol [30], stratification in 4 risk groups based on: WBC ≥ 13 × 109; BM blasts ≥ 5%; RBC transfusion dependency; Genetic risk group (includes CMML-specific cytogenetic risk stratification [23] and mutations in ASXL1, RUNX1, NRAS and SETBP1). |
|
aCML |
-Associated with the following gene mutation combinations: ASXL1/SETBP1, SETBP1/SRSF2, ASXL1/EZH2, RUNX1/EZH2 [3][4][35] |
Unfavorable outcome: mutations in TET2, RUNX1, NRAS and CUX1 [3][4] Prognostic stratification: Mayo Prognostic Model for aCML [3], stratification in 2 risk groups based on: Age > 67 years; Hb < 10 g/dL; TET2 mutations |
|
MDS/MPN-RS-T |
-WHO [1]: presence of a SF3B1 mutation. -Associated with the following gene mutation combinations: SF3B1, either alone or in combination with DNMT3A or JAK2, or DNMT3A/JAK2 [4][26][36] |
Unfavorable outcome: -Presence of altered karyotype [4][26] -Mutations in ASXL1, SETBP1, EZH2 [4][26] Prognostic stratification: Mayo Prognostic Model for MDS/MPN-RS-T [26], stratification in 3 risk groups based on: Hb < 10 g/dL; Abnormal karyotype; mutations in ASXL1 or SETBP1 |
|
MDS/MPN-U |
- |
Unfavorable outcome: -Presence of chr7 abnormalities and complex karyotypes [19] -Mutations in ASXL1, CBL, CEBPA, EZH2, STAG2, TP53 [4][27][37] Prognostic stratification: -Genomics-based stratification system (Figure 4), classification in 5 subtypes with prognostic relevance based on mutational profile [4] |
|
JMML |
-WHO [1]: presence of (1 finding sufficient): • Somatic mutation: PTPN11, KRAS, NRAS • Clinical diagnosis of NF1 or NF1 mutation • Germline CBL mutation CBL LOH |
Prognostic stratification: According to the methylation level, three groups that correlate molecular features and clinical outcome have been proposed [38]: • High: characterized by somatic PTPN11 mutations and poor clinical outcome • Intermediate: enriched in somatic KRAS mutations and monosomy 7 • Low: characterized by somatic NRAS and CBL mutations and a favorable prognosis |
4. Other Chromosomal Abnormalities
5. Functional Pathways Affected in MDS/MPN
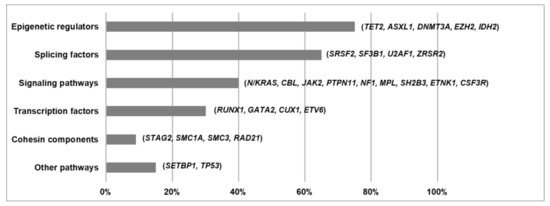
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues, 4th ed.; IARC: Lyon, France, 2017.
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Adès, L.; Fenaux, P.; Beyne-Rauzy, O.; et al. Prognostic Score Including Gene Mutations in Chronic Myelomonocytic Leukemia. JCO 2013, 31, 2428–2436.
- Patnaik, M.M.; Barraco, D.; Lasho, T.L.; Finke, C.M.; Reichard, K.; Hoversten, K.P.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Targeted next Generation Sequencing and Identification of Risk Factors in World Health Organization Defined Atypical Chronic Myeloid Leukemia. Am. J. Hematol. 2017, 92, 542–548.
- Palomo, L.; Meggendorfer, M.; Hutter, S.; Twardziok, S.; Ademà, V.; Fuhrmann, I.; Fuster-Tormo, F.; Xicoy, B.; Zamora, L.; Acha, P.; et al. Molecular Landscape and Clonal Architecture of Adult Myelodysplastic/Myeloproliferative Neoplasms. Blood 2020, 136, 1851–1862.
- Rollison, D.E.; Howlader, N.; Smith, M.T.; Strom, S.S.; Merritt, W.D.; Ries, L.A.; Edwards, B.K.; List, A.F. Epidemiology of Myelodysplastic Syndromes and Chronic Myeloproliferative Disorders in the United States, 2001–2004, Using Data from the NAACCR and SEER Programs. Blood 2008, 112, 45–52.
- Solary, E.; Itzykson, R. How I Treat Chronic Myelomonocytic Leukemia. Blood 2017, 130, 126–136.
- Patnaik, M.M.; Tefferi, A. Chronic Myelomonocytic Leukemia: 2020 Update on Diagnosis, Risk Stratification and Management. Am. J. Hematol. 2020, 95, 97–115.
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.; Sultan, C.; Cox, C. The Chronic Myeloid Leukaemias: Guidelines for Distinguishing Chronic Granulocytic, Atypical Chronic Myeloid, and Chronic Myelomonocytic Leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br. J. Haematol. 1994, 87, 746–754.
- Jaffe, E.S.; Harris, N.L.; Stein, H.; Vardiman, J.W. World Health Organization Classification of Tumours, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues; IARC Press: Lyon, France, 2001.
- Such, E.; Germing, U.; Malcovati, L.; Cervera, J.; Kuendgen, A.; Della Porta, M.G.; Nomdedeu, B.; Arenillas, L.; Luño, E.; Xicoy, B.; et al. Development and Validation of a Prognostic Scoring System for Patients with Chronic Myelomonocytic Leukemia. Blood 2013, 121, 3005–3015.
- Schuler, E.; Schroeder, M.; Neukirchen, J.; Strupp, C.; Xicoy, B.; Kündgen, A.; Hildebrandt, B.; Haas, R.; Gattermann, N.; Germing, U. Refined Medullary Blast and White Blood Cell Count Based Classification of Chronic Myelomonocytic Leukemias. Leuk. Res. 2014, 38, 1413–1419.
- Orazi, A.; Germing, U. The Myelodysplastic/Myeloproliferative Neoplasms: Myeloproliferative Diseases with Dysplastic Features. Leukemia 2008, 22, 1308–1319.
- Breccia, M.; Biondo, F.; Latagliata, R.; Carmosino, I.; Mandelli, F.; Alimena, G. Identification of Risk Factors in Atypical Chronic Myeloid Leukemia. Haematologica 2006, 91, 1566–1568.
- Giri, S.; Pathak, R.; Martin, M.G.; Bhatt, V.R. Characteristics and Survival of BCR/ABL Negative Chronic Myeloid Leukemia: A Retrospective Analysis of the Surveillance, Epidemiology and End Results Database. Ther. Adv. Hematol. 2015, 6, 308–312.
- Jeromin, S.; Haferlach, T.; Weissmann, S.; Meggendorfer, M.; Eder, C.; Nadarajah, N.; Alpermann, T.; Kohlmann, A.; Kern, W.; Haferlach, C.; et al. Refractory Anemia with Ring Sideroblasts and Marked Thrombocytosis Cases Harbor Mutations in SF3B1 or Other Spliceosome Genes Accompanied by JAK2V617F and ASXL1 Mutations. Haematologica 2015, 100, e125–e127.
- Broseus, J.; Florensa, L.; Zipperer, E.; Schnittger, S.; Malcovati, L.; Richebourg, S.; Lippert, E.; Cermak, J.; Evans, J.; Mounier, M.; et al. Clinical Features and Course of Refractory Anemia with Ring Sideroblasts Associated with Marked Thrombocytosis. Haematologica 2012, 97, 1036–1041.
- Atallah, E.; Nussenzveig, R.; Yin, C.C.; Bueso-Ramos, C.; Tam, C.; Manshouri, T.; Pierce, S.; Kantarjian, H.; Verstovsek, S. Prognostic Interaction between Thrombocytosis and JAK2 V617F Mutation in the WHO Subcategories of Myelodysplastic/Myeloproliferative Disease-Unclassifiable and Refractory Anemia with Ringed Sideroblasts and Marked Thrombocytosis. Leukemia 2008, 22, 1295–1298.
- Cannella, L.; Breccia, M.; Latagliata, R.; Frustaci, A.; Alimena, G. Clinical and Prognostic Features of Patients with Myelodysplastic/Myeloproliferative Syndrome Categorized as Unclassified (MDS/MPD-U) by WHO Classification. Leuk. Res. 2008, 32, 514–516.
- Wang, S.A.; Hasserjian, R.P.; Fox, P.S.; Rogers, H.J.; Geyer, J.T.; Chabot-Richards, D.; Weinzierl, E.; Hatem, J.; Jaso, J.; Kanagal-Shamanna, R.; et al. Atypical Chronic Myeloid Leukemia Is Clinically Distinct from Unclassifiable Myelodysplastic/Myeloproliferative Neoplasms. Blood 2014, 123, 2645–2651.
- DiNardo, C.D.; Daver, N.; Jain, N.; Pemmaraju, N.; Bueso-Ramos, C.; Yin, C.C.; Pierce, S.; Jabbour, E.; Cortes, J.E.; Kantarjian, H.M.; et al. Myelodysplastic/Myeloproliferative Neoplasms, Unclassifiable (MDS/MPN, U): Natural History and Clinical Outcome by Treatment Strategy. Leukemia 2014, 28, 958–961.
- Niemeyer, C.M.; Arico, M.; Basso, G.; Biondi, A.; Cantu Rajnoldi, A.; Creutzig, U.; Haas, O.; Harbott, J.; Hasle, H.; Kerndrup, G.; et al. Chronic Myelomonocytic Leukemia in Childhood: A Retrospective Analysis of 110 Cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood 1997, 89, 3534–3543.
- Niemeyer, C.M.; Flotho, C. Juvenile Myelomonocytic Leukemia: Who’s the Driver at the Wheel? Blood 2019, 133, 1060–1070.
- Such, E.; Cervera, J.; Costa, D.; Sole, F.; Vallespi, T.; Luno, E.; Collado, R.; Calasanz, M.J.; Hernandez-Rivas, J.M.; Cigudosa, J.C.; et al. Cytogenetic Risk Stratification in Chronic Myelomonocytic Leukemia. Haematologica 2011, 96, 375–383.
- Tang, G.; Zhang, L.; Fu, B.; Hu, J.; Lu, X.; Hu, S.; Patel, A.; Goswami, M.; Khoury, J.D.; Garcia-Manero, G.; et al. Cytogenetic Risk Stratification of 417 Patients with Chronic Myelomonocytic Leukemia from a Single Institution: Cytogenetic Stratification in CMML Patients. Am. J. Hematol. 2014, 89, 813–818.
- Wassie, E.A.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Solary, E.; Tefferi, A.; Patnaik, M.M. Molecular and Prognostic Correlates of Cytogenetic Abnormalities in Chronic Myelomonocytic Leukemia: A Mayo Clinic-French Consortium Study. Am. J. Hematol. 2014, 89, 1111–1115.
- Patnaik, M.M.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; King, R.L.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Predictors of Survival in Refractory Anemia with Ring Sideroblasts and Thrombocytosis (RARS-T) and the Role of next-Generation Sequencing: Predictors of Survival in Refractory Anemia with Ring Sideroblasts and Thrombocytosis. Am. J. Hematol. 2016, 91, 492–498.
- Mangaonkar, A.A.; Swoboda, D.M.; Coltro, G.; Lasho, T.L.; Novotny, P.J.; Pophali, P.; Carr, R.M.; Binder, M.; Finke, C.M.; Gangat, N.; et al. Clinicopathologic Characteristics, Prognostication and Treatment Outcomes for Myelodysplastic/Myeloproliferative Neoplasm, Unclassifiable (MDS/MPN-U): Mayo Clinic-Moffitt Cancer Center Study of 135 Consecutive Patients. Leukemia 2020, 34, 656–661.
- Pierre, R.V.; Hoagland, H.C. Age-Associated Aneuploidy: Loss of Y Chromosome from Human Bone Marrow Cells with Aging. Cancer 1972, 30, 889–894.
- Guttenbach, M.; Koschorz, B.; Bernthaler, U.; Grimm, T.; Schmid, M. Sex Chromosome Loss and Aging: In Situ Hybridization Studies on Human Interphase Nuclei. Am. J. Hum. Genet. 1995, 57, 1143.
- Elena, C.; Gallì, A.; Such, E.; Meggendorfer, M.; Germing, U.; Rizzo, E.; Cervera, J.; Molteni, E.; Fasan, A.; Schuler, E.; et al. Integrating Clinical Features and Genetic Lesions in the Risk Assessment of Patients with Chronic Myelomonocytic Leukemia. Blood 2016, 128, 1408–1417.
- Patnaik, M.M.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Knudson, R.A.; Ketterling, R.P.; Tefferi, A.; Solary, E. ASXL1 and SETBP1 Mutations and Their Prognostic Contribution in Chronic Myelomonocytic Leukemia: A Two-Center Study of 466 Patients. Leukemia 2014, 28, 2206–2212.
- Coltro, G.; Mangaonkar, A.A.; Lasho, T.L.; Finke, C.M.; Pophali, P.; Carr, R.; Gangat, N.; Binder, M.; Pardanani, A.; Fernandez-Zapico, M.; et al. Clinical, Molecular, and Prognostic Correlates of Number, Type, and Functional Localization of TET2 Mutations in Chronic Myelomonocytic Leukemia (CMML)-a Study of 1084 Patients. Leukemia 2020, 34, 1407–1421.
- Palomo, L.; Garcia, O.; Arnan, M.; Xicoy, B.; Fuster, F.; Cabezón, M.; Coll, R.; Ademà, V.; Grau, J.; Jiménez, M.-J.; et al. Targeted Deep Sequencing Improves Outcome Stratification in Chronic Myelomonocytic Leukemia with Low Risk Cytogenetic Features. Oncotarget 2016, 7.
- Patnaik, M.M.; Lasho, T.L.; Vijayvargiya, P.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Prognostic Interaction between ASXL1 and TET2 Mutations in Chronic Myelomonocytic Leukemia. Blood Cancer J. 2016, 6, e385.
- Meggendorfer, M.; Bacher, U.; Alpermann, T.; Haferlach, C.; Kern, W.; Gambacorti-Passerini, C.; Haferlach, T.; Schnittger, S. SETBP1 Mutations Occur in 9% of MDS/MPN and in 4% of MPN Cases and Are Strongly Associated with Atypical CML, Monosomy 7, Isochromosome i(17)(Q10), ASXL1 and CBL Mutations. Leukemia 2013, 27, 1852–1860.
- Broséus, J.; Lippert, E.; Harutyunyan, A.S.; Jeromin, S.; Zipperer, E.; Florensa, L.; Milosevic, J.D.; Haferlach, T.; Germing, U.; Luño, E.; et al. Low Rate of Calreticulin Mutations in Refractory Anaemia with Ring Sideroblasts and Marked Thrombocytosis. Leukemia 2014, 28, 1374–1376.
- Bose, P.; Nazha, A.; Komrokji, R.S.; Patel, K.P.; Pierce, S.A.; Al-Ali, N.; Sochacki, A.; Shaver, A.; Ma, W.; Su, X.; et al. Mutational Landscape of Myelodysplastic/Myeloproliferative Neoplasm-Unclassifiable. Blood 2018, 132, 2100–2103.
- Lipka, D.B.; Witte, T.; Toth, R.; Yang, J.; Wiesenfarth, M.; Nöllke, P.; Fischer, A.; Brocks, D.; Gu, Z.; Park, J.; et al. RAS-Pathway Mutation Patterns Define Epigenetic Subclasses in Juvenile Myelomonocytic Leukemia. Nat. Commun. 2017, 8, 2126.
- Sakaguchi, H.; Okuno, Y.; Muramatsu, H.; Yoshida, K.; Shiraishi, Y.; Takahashi, M.; Kon, A.; Sanada, M.; Chiba, K.; Tanaka, H.; et al. Exome Sequencing Identifies Secondary Mutations of SETBP1 and JAK3 in Juvenile Myelomonocytic Leukemia. Nat. Genet. 2013, 45, 937–941.
- Murakami, N.; Okuno, Y.; Yoshida, K.; Shiraishi, Y.; Nagae, G.; Suzuki, K.; Narita, A.; Sakaguchi, H.; Kawashima, N.; Wang, X.; et al. Integrated Molecular Profiling of Juvenile Myelomonocytic Leukemia. Blood 2018, 131, 1576–1586.
- Gondek, L.P.; Dunbar, A.J.; Szpurka, H.; McDevitt, M.A.; Maciejewski, J.P. SNP Array Karyotyping Allows for the Detection of Uniparental Disomy and Cryptic Chromosomal Abnormalities in MDS/MPD-U and MPD. PLoS ONE 2007, 2, e1225.
- Gondek, L.P.; Tiu, R.; O’Keefe, C.L.; Sekeres, M.A.; Theil, K.S.; Maciejewski, J.P. Chromosomal Lesions and Uniparental Disomy Detected by SNP Arrays in MDS, MDS/MPD, and MDS-Derived AML. Blood 2008, 111, 1534–1542.
- Tiu, R.V.; Gondek, L.P.; O’Keefe, C.L.; Elson, P.; Huh, J.; Mohamedali, A.; Kulasekararaj, A.; Advani, A.S.; Paquette, R.; List, A.F.; et al. Prognostic Impact of SNP Array Karyotyping in Myelodysplastic Syndromes and Related Myeloid Malignancies. Blood 2011, 117, 4552–4560.
- Dunbar, A.J.; Gondek, L.P.; O’Keefe, C.L.; Makishima, H.; Rataul, M.S.; Szpurka, H.; Sekeres, M.A.; Wang, X.F.; McDevitt, M.A.; Maciejewski, J.P. 250K Single Nucleotide Polymorphism Array Karyotyping Identifies Acquired Uniparental Disomy and Homozygous Mutations, Including Novel Missense Substitutions of c-Cbl, in Myeloid Malignancies. Cancer Res. 2008, 68, 10349–10357.
- Palomo, L.; Xicoy, B.; Garcia, O.; Mallo, M.; Ademà, V.; Cabezón, M.; Arnan, M.; Pomares, H.; José Larrayoz, M.; José Calasanz, M.; et al. Impact of SNP Array Karyotyping on the Diagnosis and the Outcome of Chronic Myelomonocytic Leukemia with Low Risk Cytogenetic Features or No Metaphases. Am. J. Hematol. 2016, 91, 185–192.
- Vetro, C.; Haferlach, C.; Haferlach, T.; Zenger, M.; Nadarajah, N.; Kern, W.; Meggendorfer, M. Aberrations Identified by Genomic Arrays in Normal Karyotype CMML Can Be Detected in 40% of Patients, but Do Not Add Prognostic Information to Molecular Mutations. Leukemia 2016, 30, 2235–2238.
- Jankowska, A.M.; Szpurka, H.; Tiu, R.V.; Makishima, H.; Afable, M.; Huh, J.; O’Keefe, C.L.; Ganetzky, R.; McDevitt, M.A.; Maciejewski, J.P. Loss of Heterozygosity 4q24 and TET2 Mutations Associated with Myelodysplastic/Myeloproliferative Neoplasms. Blood 2009, 113, 6403–6410.
- Kanagal-Shamanna, R.; Hodge, J.C.; Tucker, T.; Shetty, S.; Yenamandra, A.; Dixon-McIver, A.; Bryke, C.; Huxley, E.; Lennon, P.A.; Raca, G.; et al. Assessing Copy Number Aberrations and Copy Neutral Loss of Heterozygosity across the Genome as Best Practice: An Evidence Based Review of Clinical Utility from the Cancer Genomics Consortium (CGC) Working Group for Myelodysplastic Syndrome, Myelodysplastic/Myeloproliferative and Myeloproliferative Neoplasms. Cancer Genet. 2018, 228–229, 197–217.
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal Hematopoiesis of Indeterminate Potential and Its Distinction from Myelodysplastic Syndromes. Blood 2015, 9–16.
- Itzykson, R.; Kosmider, O.; Renneville, A.; Morabito, M.; Preudhomme, C.; Berthon, C.; Adès, L.; Fenaux, P.; Platzbecker, U.; Gagey, O.; et al. Clonal Architecture of Chronic Myelomonocytic Leukemias. Blood 2013, 121, 2186–2198.
- Ricci, C.; Fermo, E.; Corti, S.; Molteni, M.; Faricciotti, A.; Cortelezzi, A.; Lambertenghi Deliliers, G.; Beran, M.; Onida, F. RAS Mutations Contribute to Evolution of Chronic Myelomonocytic Leukemia to the Proliferative Variant. Clin. Cancer Res. 2010, 16, 2246–2256.
- Mason, C.C.; Khorashad, J.S.; Tantravahi, S.K.; Kelley, T.W.; Zabriskie, M.S.; Yan, D.; Pomicter, A.D.; Reynolds, K.R.; Eiring, A.M.; Kronenberg, Z.; et al. Age-Related Mutations and Chronic Myelomonocytic Leukemia. Leukemia 2016, 30, 906–913.
- Awada, H.; Nagata, Y.; Goyal, A.; Asad, M.F.; Patel, B.; Hirsch, C.M.; Kuzmanovic, T.; Guan, Y.; Przychodzen, B.P.; Aly, M.; et al. Invariant Phenotype and Molecular Association of Biallelic TET2 Mutant Myeloid Neoplasia. Blood Adv. 2019, 3, 339–349.
- Patel, B.J.; Przychodzen, B.; Thota, S.; Radivoyevitch, T.; Visconte, V.; Kuzmanovic, T.; Clemente, M.; Hirsch, C.; Morawski, A.; Souaid, R.; et al. Genomic Determinants of Chronic Myelomonocytic Leukemia. Leukemia 2017, 31, 2815–2823.




