
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sajjad Asaf | + 1171 word(s) | 1171 | 2021-05-18 11:40:37 | | | |
| 2 | Rita Xu | Meta information modification | 1171 | 2021-05-28 08:42:00 | | |
Video Upload Options
Plant secondary metabolites (SMs) play important roles in plant survival and in creating ecological connections between other species. The accumulation of SMs are highly dependent on environmental factors such as light, temperature, soil water, soil fertility, and salinity. For most plants, a change in an individual environmental factor can alter the content of secondary metabolites even if other factors remain constant. In this review, we focus on how individual environmental factors affect the accumulation of secondary metabolites in plants during both biotic and abiotic stress conditions. Furthermore, we discuss the application of abiotic and biotic elicitors in culture systems as well as their stimulating effects on the accumulation of secondary metabolites. Genes responsible for secondary metabolite biosynthesis in various plant species during stress conditions are regulated by transcriptional factors such as WRKY, MYB, AP2/ERF, bZIP, bHLH, and NAC, which are also discussed here.
1. Introduction
Compounds produced by plants are categorized into primary and secondary metabolites (SMs). Primary metabolites, such as carbohydrates, lipids, and proteins, are directly involved in plant development and growth. In contrast, SMs are multifunctional metabolites that are typically involved in plant defense and environmental communication [1]. Furthermore, they are associated with plant color, taste, and scent. Critically, they are also involved in the responses of plants to stress. For example, SMs are involved in the termination of infection, whether biotic- or abiotic-related [2]. Along with their importance in biotic stress tolerance, plant SMs are also involved in mitigating abiotic stresses such as temperature, drought, salinity, and UV light stresses [3]. When faced with certain biotic and abiotic stresses, plants can reduce morphological traits such as the number of leaves or branches, leaf area, height, and root volume [4]. Indeed, plants have a diverse array of defense mechanisms that allow them to cope with stress conditions, mitigate abiotic stress at the metabolomic level, and enhance SM accumulation during stress.Threat signals are recognized by plants’ receptors and sensors, which enable defensive responses in order to protect them from these stresses. The accumulation of secondary metabolites is one of the responses (Figure 1).Transcriptional factors (TFs) play a role in plant defense control by detecting stress signals and directing downstream defense gene expression. Similarly, the plant’s survival, durability, and productivity are all dependent on increased synthesis of secondary metabolites, known as elicitation. Various biotic (fungi, bacteria, etc.) and abiotic (exogenous hormones) elicitors are used to enhance secondary metabolites production in plants to protect them from stress stimuli (Figure 1).
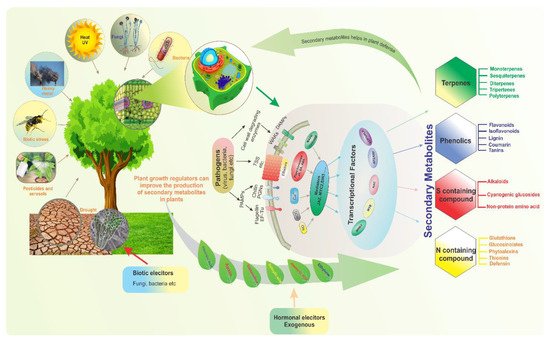
Figure 1. Diverse biotic and abiotic stresses affect plant growth and development; plants adopt various strategies and defense mechanisms to mitigate these stresses. To illustrate the modulation of secondary metabolism by various transcription factors, which are regulated by complicated upstream signaling pathways in response to stress, four plant secondary metabolite types involved in various modes of resistance are exemplified.
Plants produce SMs through several metabolic pathways that effectively respond to stress conditions. These pathways are initiated from primary metabolite pathways, which produce the ultimate precursors of SMs. The shikimate pathway is the initial pathway for biosynthesis of aromatic amino acids; it is activated in stress conditions to produced tryptophan, tyrosine, and phenylalanine, which further enhance SM biosynthesis [5]. Different SMs accumulate conditionally in various plant parts depending on the stress condition. For example, phytoalexins have antimicrobial activities against phytopathogens and accumulate at high levels in leaves [6]. In addition to their antimicrobial properties, some SMs participate in the construction of polymeric barriers to pathogen penetration.
2. Biosynthetic Pathway of SMs
The precursors of metabolites are essentially produced in the Krebs cycle and shikimate pathway. The proposed SM biosynthesis pathway is shown in Figure 2. Primary metabolites are the critical precursors of SMs. Primary and SMs can be distinguished based on their chemical structure, function, and distribution in plants. The fundamental biosynthetic pathways of metabolites are conserved in the majority of plants, with most primary metabolites found in every tissue type. The maintenance of this metabolic core has led to the occurrence of a limited number of fundamental metabolic frameworks. Frequent glycosylation, methylation, hydroxylation, acylation, oxidation, phosphorylation, and prenylation, as well as fewer chemical alterations due to tailoring of enzymes, causes a wide range of modifications in basic structures. Based on biosynthesis pathways, SMs can be divided into three main groups: phenolic compounds synthesized in the shikimate pathway, terpenes synthesized in the mevalonic pathway, and nitrogen-containing compounds synthesized in the tricarboxylic acid cycle pathway [7].
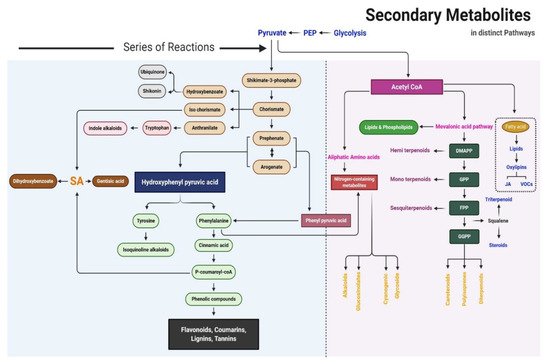
Figure 2. Schematic of the secondary metabolite biosynthesis pathway.
Shikimic acid, the precursor of the shikimate pathway, is produced from a combination of phosphoenolpyruvate (from the glycolytic pathway) and erythrose 4-phosphate (from the pentose phosphate pathway). Phenylalanine, tyrosine, and tryptophan are produced in the shikimate pathway; these are the building blocks of protein synthesis and common precursors for plant SMs such as phenolics and nitrogen-containing compounds [6]. Phenylalanine is the common precursor of flavonoids, lignans, lignins, condensed tannins, and phenylpropanoid/benzenoid volatiles; tyrosine further produces isoquinoline alkaloids, pigment betalains, and quinones (e.g., tocochromanols and plastoquinone); and tryptophan is the precursor of alkaloids, phytoalexins, indole glucosinolates, and the plant hormone auxin [8].
Seven steps are involved in the shikimate pathway to generate the end-product chorismate. In the first step, phosphoenolpyruvate and erythrose-4-phosphate are condensed to 3-deoxy-o-arabino-heptulosonate 7-phosphate (DAHP), which can formally be considered a 2-deoxy-D-glucose-6-phosphate derivative. In the second step, DAHP is exchanged with 3-dehydroquinate synthase to form the highly substituted cyclohexane derivative 3-dehydroquinate. The remaining steps in the shikimate pathway serve to add side chains and two of the three double bonds that convert this cyclohexane into a benzene ring (the hallmark of aromatic compounds). The third and fourth reaction in the shikimate pathway includes the dehydration of dehydroquinate to 3-dehydroshikimate, which is catalyzed by 3-dehydroquinate dehydratase (DHD), and the reversible reduction of 3-dehydroshikimate into shikimate using NADPH, which is catalyzed by shikimate dehydrogenase (SDH). DHD and SDH not only catalyze these respective reactions but are also part of the AROM complex in fungi. Furthermore, they are monofunctional in Escherichia coli but bifunctional in plants in the fused form of the DHD–SDH enzyme. The fifth step of the shikimate pathway is the formation of shikimate 3-phosphate. In this step, shikimate kinase catalyzes the phosphorylation of shikimate at the C3 hydroxyl group using ATP as a substrate. The sixth and seventh steps are catalyzed by 5-endolpyruvylshikimate 3-phosphate synthase (EPSP) and chorismate synthase, respectively. EPSP (also known as 3-phodphoshikimate 1-carboxyvinyltransferase) regulates the next to last step of the shikimate pathway by transferring the enopyruvyl moiety of phosphoenolpyruvate into shikimate 3-phosphate. Chorismate synthase is the final enzyme to participate in the shikimate pathway; it catalyzes the conversion of EPSP into chorismate, which is the precursor of SMs. In this final step, 1,4-anti-elimination of the 3-phosphate and C6-pro-R hydrogen from EPSP introduces the second double bond in the ring to produce chorismate. In higher plants, chorismate is the precursor of tryptophan, tyrosine, phenylalanine, salicylate, phylloquinone, and folate; it is regulated by enzymes such as chorismate mutase, iso-chorismate synthase, anthranilate synthase, and aminodeoxychorismate synthase [8].
References
- Hartmann, T. From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 2007, 68, 2831–2846.
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162.
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731.
- Pradhan, J.; Sahoo, S.; Lalotra, S.; Sarma, R. Positive impact of abiotic stress on medicinal and aromatic plants. Int. J. Plant Sci. 2017, 12, 309–313.
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737.
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90.
- Jamwal, K.; Bhattacharya, S.; Puri, S. Plant growth regulator mediated consequences of secondary metabolites in medicinal plants. J. Appl. Res. Med. Aromat. Plants 2018, 9, 26–38.
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105.




