Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivan Kushkevych | + 2369 word(s) | 2369 | 2021-05-19 11:47:58 | | | |
| 2 | Vivi Li | Meta information modification | 2369 | 2021-05-26 11:04:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kushkevych, I. Sulfate Reduction in Intestinal Bacteria. Encyclopedia. Available online: https://encyclopedia.pub/entry/10112 (accessed on 05 March 2026).
Kushkevych I. Sulfate Reduction in Intestinal Bacteria. Encyclopedia. Available at: https://encyclopedia.pub/entry/10112. Accessed March 05, 2026.
Kushkevych, Ivan. "Sulfate Reduction in Intestinal Bacteria" Encyclopedia, https://encyclopedia.pub/entry/10112 (accessed March 05, 2026).
Kushkevych, I. (2021, May 26). Sulfate Reduction in Intestinal Bacteria. In Encyclopedia. https://encyclopedia.pub/entry/10112
Kushkevych, Ivan. "Sulfate Reduction in Intestinal Bacteria." Encyclopedia. Web. 26 May, 2021.
Copy Citation
Sulfate is present in foods, beverages, and drinking water. Its reduction and concentration in the gut depend on the intestinal microbiome activity, especially sulfate-reducing bacteria (SRB), which can be involved in inflammatory bowel disease (IBD). Assimilatory sulfate reduction (ASR) is present in all living organisms. In this process, sulfate is reduced to hydrogen sulfide and then included in cysteine and methionine biosynthesis. In contrast to assimilatory sulfate reduction, the dissimilatory process is typical for SRB. A terminal product of this metabolism pathway is hydrogen sulfide, which can be involved in gut inflammation and also causes problems in industries (due to corrosion effects).
intestinal microbiota
sulfate reduction
assimilatory
sulfate-reducing bacteria
hydrogen sulfide
toxicity
cysteine biosynthesis
1. Introduction
Sulfur is an indispensable element for living organisms, including prokaryotes and eukaryotes. This element is important in the synthesis of amino acids, proteins, and enzymes [1]. Sulfur metabolism depends on different organisms and the environment. These factors determine its oxidation state. Microorganisms can involve sulfur in their metabolism, both in oxidized and reduced states [2].
The intestinal microorganisms utilize mostly the oxidized state of sulfur from various complex organic compounds (such as sulfate present in food and water). The intestinal microbiome has a key impact on human health and can be involved in immunity, metabolism, and neurobehavioral traits. Although intestinal bacteria have been studied for several decades, their role in the intestines has been found more interesting than classical infectious microorganisms [3].
The reduction of sulfate, depending on microorganism, can take place in two ways: assimilatory and dissimilatory (differing in the final product) [1][2]. In dissimilatory sulfate reduction (DSR), toxic hydrogen sulfide is produced, and in an assimilatory way, the terminal product is cysteine. One possible hypothesis of intestinal inflammation, including ulcerative colitis, is the effect of high concentrations of hydrogen sulfide on the intestinal epithelium [4][5][6][7][8][9][10]. In recent years, intestinal sulfate-reducing bacteria (SRB) (as a common part of the intestinal microbiome) have been associated with this disease [11][12][13].
SRB are anaerobic bacteria that use organic compounds as a source of energy and carbon in a process called DSR [14][15][16][17]. This is typical of only five bacterial strains [18]. In this metabolic pathway, sulfate is used as an electron acceptor and is reduced to hydrogen sulfide, which is subsequently released into the intestine [4][19][20][21]. SRB are not the only bacteria that use sulfate and produce hydrogen sulfide. There are some other bacteria in the intestine, such as Escherichia coli, that metabolize sulfate to hydrogen sulfide, and further to sulfur amino acids. This metabolic pathway is called assimilatory sulfate reduction (ASR) and occurs in both bacteria and plants [22].
2. Sulfate Source for Intestinal Bacteria
The sulfate contained in food plays an important role in human metabolism. It is involved in the formation of methionine and cysteine. In conjugation with xenobiotics and drugs, sulfate influences the metabolism of the large intestine, where it is reduced by bacteria to toxic and harmful intestine hydrogen sulfide [12][13]. However, it is poorly absorbed in the intestine and can be used as a laxative or contrast agent, such as Ba2SO4. An adult human body receives more than 16 mmol of sulfate per day [23]. A portion of the sulfate is absorbed in the small intestine and is used to form sulfur-containing compounds, such as chondroitin sulfate and mucin, or is used for the synthesis of amino acids methionine and cysteine [23]. Cysteine is an indispensable component of peptides, involved in the formation of disulfide bridges, and is highly represented in keratin protein. Keratin protein can be found in hair and nails, and it is included in the formation of taurine. Cysteine decarboxylation produces mercaptoethanolamine, which is involved in the biosynthesis of coenzyme A [24]. The remaining sulfate ions that are not absorbed in the small intestine, reach the colon, where they serve as electron acceptors for SRB [4]. The largest sources of sulfate in the diet are industrially processed foods such as wheat bread (15 μmol/g), sausages (10 μmol/g), but also nuts (9 μmol/g) and dried fruit (10 μmol/g). A high sulfate content occurs in beer (2.6 μmol/mL) and some vegetables (8.4 μmol/g); mainly vegetables, the most often from the cabbage family, such as cauliflower, which is a rich source of glucosinolates. Their general formula is R-C-(NOSO3)-S-glucose [23]. Besides sulfate, there are other sulfur compounds in the intestine. Other intestinal bacteria are involved in the processing of these sulfur compounds (Table 1), which are necessary to maintain a balance between the harmful and beneficial effects of sulfur-containing compounds on the intestinal tract [2].
3. Dissimilatory Sulfate Reduction
Only a special group of organisms can extract energy from DSR, while the ASR process is abundant among other organisms (plants and bacteria). SRB are an important group of microorganisms in various ecosystems, including the intestinal tract, because they require inorganic sulfate as an electron acceptor to obtain energy from organic compounds [25][26]. SRB are a significant part of the environment because they require inorganic sulfate for energy production [18]. DSR is a process that includes several reactions, providing energy for the creation of adenosine triphosphate (ATP) in bacterial cells of SRB.
Table 1. Various bacterial genera involved in sulfur metabolism (modified from Carbonero et al. 2012) [2].
| Source of Sulfate | Substrates Containing Sulfur | Bacteria Genera | Reference |
|---|---|---|---|
| Inorganic | Sulfate (SO42−) | Desulfovibrio sp. | Gibson et al. 1988 [12] |
| Desulfobacter sp. | |||
| Desulfobulbus sp. | |||
| Desulfotomaculum sp. | |||
| Sulfite (SO32−) | Bilophila wadsworthia | Baron et al. 1989 [27] | |
| Campylobacter jejuni | Kelly and Myers 2005 [28] | ||
| Organic | Cysteine | Escherichia coli | Metaxas and Delwiche 1955 [29] |
| Staphylococcus aureus | Shatalin et al. 2011 [30] | ||
| Salmonella typhimurium | Kredich et al. 1972 [31] | ||
| Mycobacterium tuberculosis | Wheeler et al. 2005 [32] | ||
| Helicobacter pylori | Kim et al. 2006 [33] | ||
| Prevotella intermedia | Igarashi et al. 2009 [34] | ||
| Fusobacterium nucleatum | Yoshida et al. 2010 [35] | ||
| Streptococcus anginosus | Yoshida et al. 2010 [36] | ||
| Clostridium sp. | Genom cysteine desulfohydrase | ||
| Enterobacter sp. | Genom cysteine desulfohydrase | ||
| Klebsiella sp. | Genom cysteine desulfohydrase | ||
| Sulfomucin | Prevotella strain RS2 | Roberton et al. 2000 [37] | |
| Bacteroides fragilis | Roberton et al. 2000 [37] | ||
| Helicobacter pylori | Slomiany et al. 1992 [38] | ||
| Akkermansia sp. | Genom glycosulfatase |
Sulfate is used as a terminal acceptor and provides energy for growth and is released from the oxidation of organic compounds (lactate, propionate, and butyrate) or hydrogen [39][40]. Reduction of sulfate occurs over many different intermediates that are not released to the environment. The process of DSR includes the transfer of eight electrons [4]. Enzymes that are associated with this process are localized in the cytoplasm or periplasm of the bacterial cells. The first step is sulfate and proton absorption into bacterial cells [41][42][43][44][45]. After these processes, DSR can be divided into six stages (Figure 1).
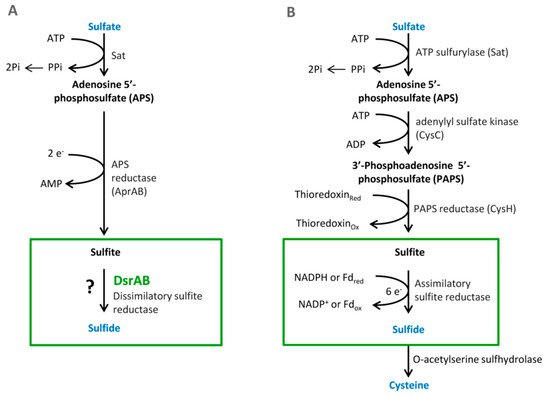
Figure 1. The pathways of the dissimilatory (A) and assimilatory (B) sulfate reduction, from Santos et al. 2015 [46].
3.1. Sulfate Activation
Sulfate must be reduced and activated before absorption. This enzymatic reaction is catalyzed by the enzyme ATP sulfurylase (EC 2.7.7.4.). The products of this reaction are adenosine-5-phosphosulfate (APS) and pyrophosphate (PPi) [14]. Firstly, free sulfate around bacterial cells must be activated by APS sulfurylase that binds ATP to sulfate. This enzyme is part of three metabolic pathways: purine, selenium, and sulfur metabolism. APS sulfurylase is also found in plants, where it is involved in sulfur metabolism as well [4]. In the bacterium Desulfovibrio vulgaris Hildenborough, APS sulfurylase is encoded by the sat gene. This gene is localized on the negative strand in the position 1,387,712–1,388,995. It is composed of 184 nucleotides, which are translated into 427 amino acids. The active site consists of 142 amino acids at position 209–350. The polypeptide chain contains two protein motives, a flexible turn and an HXXH motive. The HXXH motive participates in catalytic reactions. These motives are typical for proteins from the nucleotidyltransferase protein family [47].
3.2. Cytoplasmic APS Reduction
This reaction is catalyzed by the enzyme APS reductase (EC 1.8.99.2), which enables the reduction of APS to sulfite or bisulfite and adenosine monophosphate (AMP) [42][43]. In the bacterium D. vulgaris, sulfite exists only as an intermediate [28]. This reaction is catalyzed by the enzyme APS reductase. It can be found in SRB and also in other sulfuric bacteria or in Thiobacillus. APS reductase is a nonheme flavoprotein that consists of two subunits: α and β [44]. In the genome of Desufovibrio vulgaris, Hildenborough, genes encode both subunits, and they are situated in a positive operon strand (Figure 2) [47].
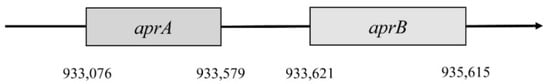
Figure 2. Localization of aprA and aprB genes.
The first gene encodes subunit β. It is named aprB and is situated in the 933,076 to 933,579 position. This gene consists of 504 nucleotides that are translated into 167 amino acids. Subunit β has two domains 4Fe–4S; these are encoded by 49 amino acids, forming a place for cofactors. Gene aprA encodes subunit α; this gene is in position 933,621–935,165. It consists of 1995 nucleotides, which are translated into 644 amino acids. This subunit reacts with flavin adenine dinucleotide (FAD) [4][47]. The gene for subunit α is almost four times larger than the aprB gene. Both genes must be correctly translated into proteins, and then they bind with FAD and iron. This complex is a fully functional holoenzyme that is called APS-reductase [4].
3.3. Cytoplasmic Sulfite Reduction
Another essential reaction of the process is sulfite reduction, resulting in the product of APS reduction [4][44]. Sulfite reduction is catalyzed by dissimilatory sulfite reductase (EC 1.8.99.1). This enzyme reduces sulfite to sulfate [44]. Sulfide reductase also plays an important role in the process of assimilatory sulfate reduction due to sulfide ion production. These sulfides are part of amino acids containing sulfur, such as methionine and cysteine. SRB may have several types of sulfide reductases that can be used for identification. These reductases are desulfoviridin, desulforubin, desulfofuscidine, and protein P582 [4].
Sulfite reduction is the last reaction in the process of DSR. Reactive sulfite is converted into toxic sulfide, and then it is released out of the bacterial cell [48]. This reaction is catalyzed by the enzyme sulfite reductase (Figure 3).
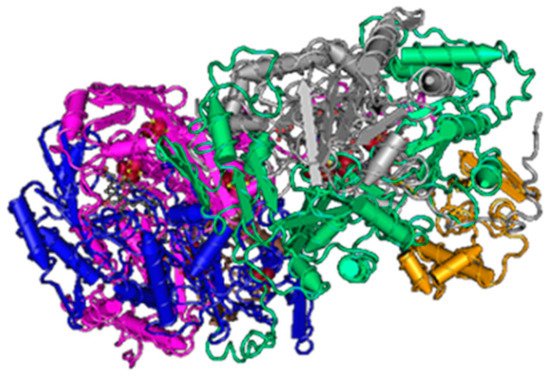
Figure 3. The structure of sulfite reductase (3D structure) [46].
Various species of SRB have different types of this enzyme (desulfoviridin, desulforubidin, desulfofuscidine, P582). The best-known type is desulfovidirin that can be found in the genus Desulfovibrio. In 2010, Barton and Hamilton discovered that this enzyme consists of 3 subunits α, β, γ. They proved the existence of the hexamer structure 2α2β2γ [44]. Every single subunit is encoded by its own gene, but they are all located in the positive strand of the chromosome. Desulforubidin is typical for genera Desulfohalobium, Desulfosarcina, Desulfomicrobium, Desulfocurvus, Desulfobulbus, Desulfofustis, and Desulfobacter. Desulforubidin has a very similar structure to desulfoviridin [1]. Desulfofuscidin consists of only two subunits, α and β and has been isolated from thermophilic SRB (Thermodesulfovibrio) [49][50][51].
In Desulfovibrio vulgaris, Hildenborough, subunit α is encoded by the gene dsvA. This gene is in the position 449,888–451,201. It consists of 1314 nucleotides that are translated into a protein with a length of 437 amino acids, and part of this protein is also the domain 4Fe–4S with a length of 231 amino acids [4]. This part belongs to the protein family Nir-Sir. Next to gene dsvA is gene dvsB that encodes subunit β. This gene is in position 451,220–452,365 and contains 1146 nucleotides that are translated into a protein with a length of 222 amino acids. The last gene called dsvC is responsible for the subunit γ that is located in a different part of the chromosome (position 2,883,462–2,883,779). This protein consists of 105 amino acids [47].
Genes dsvA and dvsB are translated as operons (Figure 4), and they form a tetramer structure from subunits α and β, and thus they comprise the center of sulfite reductase.
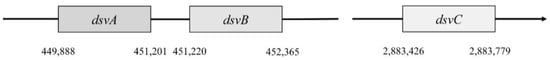
Figure 4. Localization of dsvA, dsvB, dsvC genes.
3.4. Periplasmic Oxidation of Molecular Hydrogen
Oxidation of H2 occurs in the periplasm by periplasmic dehydrogenases. These hydrogenases are enzymes that catalyze reversible reactions in the presence of hydrogen. They are crucial for anaerobic respiration.
3.5. Transmembrane Transfer of Electrons
This process takes place in the periplasm as well. Protons are transferred to cytochrome c3 by periplasmic dehydrogenase [44].
3.6. Cytoplasmic Oxidation of Molecular Hydrogen
This reaction is catalyzed by cytoplasmic hydrogenases and FeS proteins.
The first three enzymatic reactions are important for sulfate reduction to hydrogen sulfide (Figure 5). Enzymes of mentioned reactions are connected directly with inorganic sulfate and they are typical for the DSR pathway. The next three reactions are linked to electron transport.
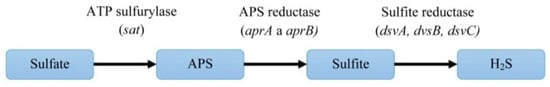
Figure 5. The pathway of dissimilatory sulfate reduction and the genes encoding the enzymes of this process.
4. The Relationship of Sulfate Reduction Pathways and Inflammatory Bowel Disease (IBD)
Sulfate consumption and its conversion to hydrogen sulfide affect the pH of the intestines. The colon is an unfavorable environment for SRB due to pH lower than 5.5. By contrast, the distal part of the colon has neutral pH and is an optimal environment for SRB. The acidic pH of the intestines influences the occurrence of inflammatory bowel diseases (IBD) [4][9].
Ulcerative proctitis occurs in the most distal part of intestine and the rectum, distal colitis in the descending colon, and pancolitis in the entire colon [53][54]. The reason for the occurrence of ulcerative colitis and IBD has not been fully revealed yet. Factors, such as dietary habits and intestinal microbiome composition, influence IBD prevalence [26]. A connection between the SRB intestinal occurrence and IBD has been found. However, these bacteria are still considered an ordinary component of the digestive system of healthy people. It was calculated that the healthy population has SRB prevalence in the digestive system from 12% to 79% [11][12].
SRB can be considered only a contributing factor to ulcerative colitis development since they are not direct pathogens [12]. The main role of SRB in IBD can be explained by their production of hydrogen sulfide due to inhibition of butyrate oxidation. Without butyrate oxidation, colon cells starve since they cannot oxidase short chain fatty acids (mainly butyrate). On the other hand, apoptosis of colonic cancer cells is supported by short chain fatty acids [53]. Lower butyrate oxidation levels were observed in studies that included UC patients. The normal concentration of hydrogen sulfide in the gut ranges from 1.0 mM to 2.4 mM. These concentrations are even considered beneficial for the colonic mucosa (cell respiration is increased) [54].
Food and beverages are sources of sulfate; approximate daily intake is from 2 mmol to 9 mmol. These amounts are mainly utilized and only around 0.5 mmol is measured in daily fecal matter. The complexity of the gut microbiota can also be seen by symbiotic processes between SRB and saccharolytic bacteria. Saccharolytic bacteria disengage bound sulfate [11].
SRB influence the prevalence of IBD, but they are certainly not the only factor. The production of hydrogen sulfide affects the colon environment, but excessive amounts of hydrogen sulfide are also toxic for SRB [5].
As already mentioned, hydrogen sulfide is the final product in the metabolism of SRB in the dissimilatory sulfate reduction process. SRB represent the main hazardous issues considering the occurrence of SRB in the gut microbiota. Thus, the pathway switch from DSR to ASR can be seen as the opportunity to decrease hydrogen sulfide formation by intestinal SRB or at least its production in lower quantities.
References
- Barton, L.L.; Fauque, G.D. Biochemistry, physiology and biotechnology of sulfate-reducing bacteria. Adv. Appl. Microbiol. 2009, 68, 41–98.
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448.
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725.
- Kushkevych, I. Dissimilatory sulfate reduction in the intestinal sulfate-reducing bacteria. Studia Biologica 2016, 10, 197–228.
- Kushkevych, I.; Dordević, D.; Vítězová, M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria Desulfovibrio piger Vib-7. Arch. Microbiol. 2019, 201, 389–397.
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen Sulfide as a Toxic Product in the Small–Large Intestine Axis and its Role in IBD Development. J. Clin. Med. 2019, 8, 1054.
- Kushkevych, I.; Kotrsová, V.; Dordević, D.; Buňková, L.; Vítězová, M.; Amedei, A. Hydrogen Sulfide Effects on the Survival of Lactobacilli with Emphasis on the Development of Inflammatory Bowel Diseases. Biomolecules 2019, 9, 752.
- Kotrsová, V.; Kushkevych, I. Possible methods for evaluation of hydrogen sulfide toxicity against lactic acid bacteria. Biointerface Res. Appl. Chem. 2019, 9, 4066–4069.
- Kushkevych, I.; Leščanová, O.; Dordević, D.; Jančíková, S.; Hošek, J.; Vítězová, M.; Buňková, L.; Drago, L. The Sulfate-Reducing Microbial Communities and Meta-Analysis of Their Occurrence during Diseases of Small–Large Intestine Axis. J. Clin. Med. 2019, 8, 1656.
- Barton, L.L.; Fardeau, M.L.; Fauque, G.D. Hydrogen sulfide: A toxic gas produced by dissimilatory sulfate and sulfur reduction and consumed by microbial oxidation. Met. Ions Life Sci. 2014, 14, 237–277.
- Gibson, G.R.; Macfarlane, S.; Macfarlane, G.T. Metabolic interactions involving sulfate-reducing and methanogenic bacteria in the human large intestine. FEMS Microbiol. Ecol. 1993, 12, 117–125.
- Gibson, G.R.; Macfarlane, G.T.; Cummings, J.H. Occurrence of sulfate-reducing bacteria in human faeces and the relationship of dissimilatory sulfate reduction to methanogenesis in the large gut. J. Appl. Bacteriol. 1988, 65, 103–111.
- Gibson, G.R. Physiology and ecology of the sulfate-reducing bacteria. J. Appl. Bacteriol. 1990, 69, 769–797.
- Kushkevych, I.; Fafula, R.; Parak, T.; Bartoš, M. Activity of Na+/K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Acta Vet. Brno. 2015, 84, 3–12.
- Kushkevych, I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Pol. J. Microbiol. 2015, 64, 107–114.
- Kushkevych, I.; Vítězová, M.; Kos, J.; Kollár, P.; Jampilek, J. Effect of selected 8-hydroxyquinoline- 2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J. Appl. Biomed. 2018, 16, 241–246.
- Kushkevych, I.; Kollar, P.; Suchy, P.; Parak, T.; Pauk, K.; Imramovsky, A. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuro Endocrinol. Lett. 2015, 36, 106–113.
- Wagner, M.; Roger, A.J.; Flax, J.L.; Brusseau, G.A.; Stahl, D.A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 1998, 180, 2975–2982.
- Kushkevych, I.V. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochem. Pol. 2015, 62, 1037–1108.
- Kushkevych, I.; Dordević, D.; Vítězová, M.; Kollár, P. Cross-correlation analysis of the Desulfovibrio growth parameters of intestinal species isolated from people with colitis. Biologia 2018, 73, 1137–1143.
- Kushkevych, I.; Dordević, D.; Vítězová, M. Analysis of pH dose-dependent growth of sulfate-reducing bacteria. Open Med. (Wars.) 2019, 14, 66–74.
- Schiff, J.A. Pathways of assimilatory sulphate reduction in plants and microorganisms. Ciba Found. Symp. 1979, 72, 49–69.
- Florin, T.H.J.; Neale, G.; Goretski, S.; Cumming, J.H. The sulfate content of foods and beverages. J. Food Compos. Anal. 1993, 6, 140–151.
- Weinstein, C.L.; Haschemeyer, R.H.; Griffith, O.W. In vivo studies of cysteine metabolism. Use of D-cysteinesulfinate, a novel cysteinesulfinate decarboxylase inhibitor, to probe taurine and pyruvate synthesis. J. Biol. Chem. 1988, 263, 16568–16579.
- Kováč, J.; Vítězová, M.; Kushkevych, I. Metabolic activity of sulfate-reducing bacteria from rodents with colitis. Open Med. (Wars.) 2018, 13, 344–349.
- Kushkevych, I.; Vítězová, M.; Fedrová, P.; Vochyanová, Z.; Paráková, L.; Hošek, J. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet. Brno 2017, 86, 405–411.
- Baron, E.J.; Summanen, P.; Downes, J.; Roberts, M.C.; Wexler, H.; Finegold, S.M. Bilophila wadsworthia, gen-nov and sp-nov, a unique Gram-negative Anaerobic rod recovered from appendicitis specimens and human feces. J. Gen. Microbiol. 1989, 135, 3405–3411.
- Kelly, D.J.; Myers, J.D. A sulfite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 2005, 151, 233–242.
- Metaxas, M.A.; Delwiche, E.A. The L-cysteine desulfhydrase of Escherichia coli. J. Bacteriol. 1955, 70, 735–737.
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 2011, 334, 986–990.
- Kredich, N.M.; Keenan, B.S.; Foote, L.J. Purification and subunits structure of cysteine desulfhydrase from Salmonella typhimurium. J. Biol. Chem. 1972, 247, 7157–7162.
- Wheeler, P.R.; Coldham, N.G.; Keating, L.; Gordon, S.V.; Wooff, E.E.; Parish, T.; Hewinson, R.G. Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 2005, 280, 8069–8078.
- Kim, Y.K.; Lee, H.; Kho, H.S.; Chung, J.W.; Chung, S.C. Volatile sulfur compounds produced by Helicobacter pylori. J. Clin. Gastroenterol. 2006, 40, 421–426.
- Yano, T.; Fukamachi, H.; Yamamoto, M.; Igarashi, T. Characterization of L-cysteine desulfhydrase from Prevotella intermedia. Oral Microbiol. Immunol. 2009, 24, 485–492.
- Yoshida, Y.; Ito, S.; Kamo, M.; Kezuka, Y.; Tamura, H.; Kunimatsu, K.; Kato, H. Production of hydrogen sulfide by two enzymes associated with biosynthesis of homocysteine and lanthionine in Fusobacterium nucleatum subsp. pnucleatum ATCC25586. Microbiology 2010, 156, 2260–2269.
- Yoshida, A.; Takahashi, Y.; Nagata, E.; Hoshino, T.; Oho, T.; Awano, S.; Takehara, T.; Ansai, T. Streptococcus anginosusl cysteine desulfhydrase gene expression is associated with abscess formationin BALB/c mice. Mol. Oral Microbiol. 2011, 26, 221–227.
- Wright, D.P.; Rosendale, D.I.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79.
- Slomiany, B.L.; Murty, V.L.N.; Piotrowski, J.; Grabska, M.; Slomiany, A. Glycosulfatase activity of Helicobacter pylori toward human gastric mucin effect of sucralfate. Am. J. Gastroenterol. 1992, 87, 1132–1137.
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017, 12, 82–91.
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Kováč, J.; Kaucká, P.; Jesionek, W.; Bartoš, M.; Barton, L. A new combination of substrates: Biogas production and diversity of the methanogenic microorganisms. Open Life Sci. 2018, 13, 119–128.
- Kushkevych, I.; Dordević, D.; Kollar, P. Analysis of physiological parameters of Desulfovibrio strains from individuals with colitis. Open Life Sci. 2018, 13, 481–488.
- Friedrich, M.W. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5’-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 2002, 184, 278–289.
- Rabus, R.T.; Hansen, A.; Widdel, F. Dissimilatory Sulfate and Sulfur-Reducing Prokaryotes. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackenbrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 659–768.
- Barton, L.L.; Hamilton, W.A. Sulfate-Reducing Bacteria: Environmental and Engineered Systems; Cambridge University Press: Cambrige, UK, 2010; 553p.
- Michaels, G.B.; Davidson, J.T.; Peck, H.D., Jr. A flavin-sulfite adduct as an intermediate in the reaction catalyzed by adenylyl sulfate reductase from Desulfovibrio vulgaris. Biochem. Biophys. Res. Commun. 1970, 39, 321–328.
- Santos, A.A.; Venceslau, S.S.; Grein, F.; Leavitt, W.D.; Dahl, C.; Johnston, D.T.; Pereira, I.A. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 2015, 350, 1541–1545.
- Heidelberg, J.F.; Seshadri, R.; Haveman, S.A.; Hemme, C.L.; Paulsen, I.T.; Kolonay, J.F.; Eisen, J.A.; Ward, N.; Methe, B.; Brinkac, L.M.; et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 2004, 22, 554–559.
- Oliveira, T.F.; Vonrhein, C.; Matias, P.M.; Venceslau, S.S.; Pereira, I.A.; Archer, M. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J. Biol. Chem. 2008, 283, 34141–34149.
- Fauque, G.; Lino, A.R.; Czechowski, M.; Kang, L.; Der Vartanian, D.V.; Moura, J.J.; LeGall, J.; Moura, I. Purification and characteriztion of bisulfite reductase (desulfofuscidin) from Desulfovibrio thermophilus and its complexes with exogenous ligands. Biochem. Biophys. Acta 1990, 1040, 112–118.
- Kushkevych, I.; Cejnar, J.; Vítězová, M.; Vítěz, T.; Dordević, D.; Bomble, Y.J. Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants. Int. J. Mol. Sci 2020, 21, 283.
- Fauque, G.; Le Gall, J.; Barton, L.L. Sulfur reductase from thiophilic sulfate-reducing bacteria. In Variations in Autotrophic Life; Shively, J.M., Barton, L.L., Eds.; Academic Press: London, UK, 1991; pp. 271–337.
- Karkhoff-Schweizer, R.R.; Huber, D.W.; Voordouw, G. Conservation of the Genes for Dissimilatory Sulfite Reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus Allows Their Detection by PCR. Appl. Environ. Microbiol. 1995, 61, 290–296.
- Rowan, F.E.; Docherty, N.G.; Coffey, J.C.; O’Connell, P.R. Sulphate-reducing bacteria and hydrogen sulphide in the etiology of ulcerative colitis. Br. J. Surg. 2009, 96, 151–158.
- Roediger, W.E. The colonic epithelium in ulcerative colitis: An energy-deficiency disease? Lancet 1980, 2, 712–715.
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
26 May 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





