Video Upload Options
Kinases are considered the largest protein family in the human proteome, and approximately 2% of eukaryotic genes encode kinase superfamily members. These enzymes catalyze the transference of the γ-phosphate from ATP to serine, threonine, or tyrosine amino acid residues of a downstream protein substrate, creating a communication cascade that is fundamental to eukaryotic cells.
1. General introduction
Over 500 kinases were identified in humans. The so-called ‘human kinome’ is divided into typical kinases and atypical kinases. The 478 typical kinases possess a well-defined architecture that is similar across all members, while the 40 atypical kinases (the ‘dark kinome’) are poorly understood [1]. Most typical kinases are dually phosphorylated at serine and threonine residues and, as a result, are called Ser/Thr kinases. Kinases are clustered according to similarities at the kinase domain, which results in different kinase groups/families. According to evolutionary conservation data, the CMGC kinase group is an ancient group made up by nine highly conserved families found in most eukaryotes [2]. This group was named after the initials of some of its key members: cyclin-dependent kinase (CDK), MAPK, glycogen synthase kinase (GSK), and CDK-like kinases (CLK).
2. The MAPK family
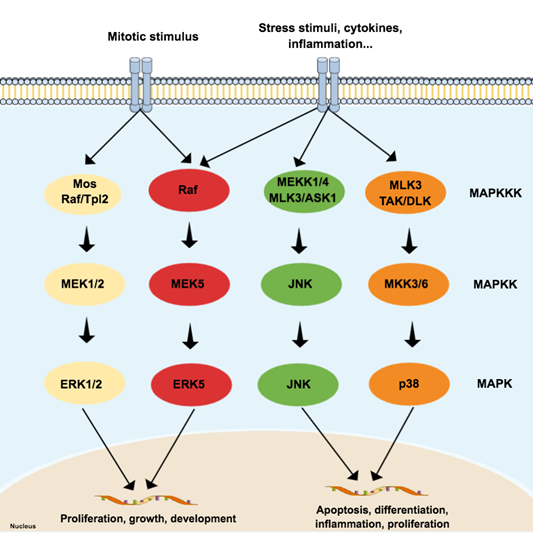
The MAPK family is considered the main propagator of extracellular signals from the cell membrane to the nucleus as they catalyze the transfer of the γ-phosphate from ATP to serine or threonine residues of various substrates [3], including transcription factors, which regulate the expression of specific sets of genes, and thus mediate a specific response according to the stimulus received by cells [4][5][6][7]. Among the MAPK family, fourteen members share both structures and biochemical properties. Depending on those characteristics, each member is grouped in one of the seven different MAPK subfamilies. The extracellular signal-regulated kinases 1/2 (ERK1/2), ERK5, JNK, and p38 subfamilies can be activated by dual phosphorylation of threonine and tyrosine residues (known as ‘tripeptide motif’ or ‘Thr-X-Tyr motif’) at the activation loop site (also called ‘A-loop’) and activate the pathway by propagating the phosphorylation in downstream kinases. Those four subfamilies follow a classical three-tiered signaling pathway that is highly conserved in eukaryotic organisms, where each phosphorylation of a MAPK is carried out by specific upstream kinases. Usually, MAPKKK (MAP3K) receives a variety of inputs and transmits them to downstream kinases MAPKK (MAP2K), which activates MAPK (Figure 1) [8][9][10]. On the other hand, ERK3/4, ERK7/8, and NLK (nemo-like kinase) compose the ‘atypical’ MAPK subfamilies that do not follow the same dual-phosphorylation and three-tiered module pattern. However, since the focus of this paper is on ‘typical’ MAPK—specifically in JNK—the ‘atypical’ MAPK will not be reviewed in detail and will be referred to as appropriate. A detailed review of their dynamics can be checked elsewhere [8][9].
Figure 1. Schematic representation of the typical mitogen-activated protein kinases (MAPK) pathway, composed of three levels (three-tier module) and four subfamilies: extracellular signal-regulated kinases (ERK) 1/2, p38, c-Jun N-terminal kinase (JNK), and ERK5. Each subfamily regulates different phenotypes. While the canonical ERK subfamily (ERK1/2) responds to mitotic stimuli and controls cellular differentiation and proliferation, JNK and p38 are activated by stressor stimuli and are involved in apoptosis. Since the ERK5 subfamily responds to both mitotic and stressor stimuli, being associated with cell survival, it is presented in a special subfamily.
MAP3K activates more than one MAP2K, and MAP2K can phosphorylate more than one MAPK. The cross-talking is important to signal integration and coordination but may also represent an obstacle in therapy since it is likely to be involved in drug resistance in cancer, for example [11]. Although the MAPK pathways have several interconnections that sometimes hinder the separation of these pathways, the JNK and p38 pathways are both activated by cellular stressors, such as cytokines for example, and are frequently associated with cellular death and inflammation [4][7][12]. On the other hand, both JNK and p38 can mediate anti-apoptotic events as well and do not always work ‘as a team’, since recent evidence suggests negative regulation of p38 over JNK in some cellular contexts [13].
3. JNK
JNK is a MAPK activated by pro-inflammatory cytokines or exposure to environmental stress and is best known for its role in programed cell death [14]. The term ‘apoptosis’ was suggested in 1972, and it describes a regulated cell death process that requires a complex molecular program of self-destruction where cells retain plasma integrity and some level of metabolic activity until the outcome [15]. Apoptotic processes are divided into extrinsic and intrinsic pathways, and a detailed review of this topic is available elsewhere [15][16]. Here we will discuss general aspects that involve the role of JNK in apoptosis, as illustrated in Figure 2.
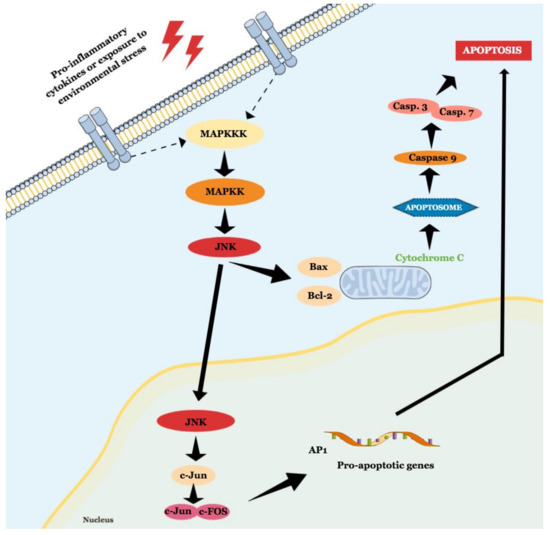
Figure 2. Schematic representation of JNK role in intrinsic and extrinsic apoptosis.
The intrinsic pathway is activated by extracellular or intracellular perturbations usually found in AD, such as oxidative stress and microtubular alterations caused by NTFs [17][18]. This pathway is controlled by members of the Bcl-2 family, which is divided into three groups known as pro-apoptotic pore-formers (Bax, Bak, and Bok), pro-apoptotic BH3-only proteins (Bad, Bid, Bik, Bmf, Hrk, Noxa, Puma, etc), and anti-apoptotic proteins (Bcl-2, Bcl-XL, Bcl-W, Mcl-1, Bfl-1) [19]. The Bcl-2 family is under the control of JNK and p38 [13], and a significant part of them can transit between cytosol and organelles. In response to a deleterious stimulus, the JNK-mediated phosphorylation of 14-3-3 protein at the Ser184/186 site induces the translocation of pro-apoptotic proteins (Bax and Bad) from the cytoplasm to the mitochondria. However, it was reported that JNK can directly phosphorylate Bad at Ser128, Bim at Ser65, and Bid at Thr59, inducing the pro-apoptotic activity, and inhibiting anti-apoptotic proteins, since Bad, Bim, and Bmf inhibit Bcl-2 antiapoptotic effect [16][18][20]. JNK also phosphorylates Bcl-2 and Mcl-1, blocking their anti-apoptotic activity. This pathway is also known as the ‘mitochondrial pathway’ since the relocation of pro-apoptotic proteins causes mitochondrial outer membrane permeabilization (MOMP), allowing the cytosolic release of pro-apoptogenic factors that normally reside in the mitochondrial intermembrane space, such as cytochrome c and Smac/DIABLO. Cytochrome c then associates with Apaf-1, pro-caspase 9 (CASP9), (and possibly other proteins) to form an apoptosome, which activates CASP9. When activated, CASP9 catalyzes the proteolytic activation of CASP3 and CASP7 (known as ‘executioner caspases’), which handle cell demolition during intrinsic and extrinsic apoptosis pathways. In this sense, both pathways can induce caspase activation that causes the morphological and biochemical features commonly seen during apoptosis, including DNA fragmentation and phosphatidylserine exposure [21]. However, both the pathways usually operate independently, since Bid, a pro-apoptotic Bcl-2 family member, can be cleaved by CASP8, promoting cytochrome c release [18].
References
- Modi, V.; Dunbrack, R.L. A Structurally-Validated Multiple Sequence Alignment of 497 Human Protein Kinase Domains. Sci. Rep. 2019, 9, 19790.
- Manning, G. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934.
- Pei, J.J.; Braak, E.; Braak, H.; Grundke-Iqbal, I.; Iqbal, K.; Winblad, B.; Cowburn, R.F. Localization of active forms of C-jun kinase (JNK) and p38 kinase in Alzheimer’s disease brains at different stages of neurofibrillary degeneration. J. Alzheimer’s Dis. 2001, 3, 41–48.
- Kanungo, J. DNA-PK and P38 MAPK: A Kinase Collusion in Alzheimer’s Disease? Brain Disord. Ther. 2017.
- Borroto-Escuela, D.O.; Tarakanov, A.O.; Guidolin, D.; Ciruela, F.; Agnati, L.F.; Fuxe, K. Moonlighting characteristics of G protein-coupled receptors: Focus on receptor heteromers and relevance for neurodegeneration. IUBMB Life 2011, 63, 463–472.
- Shi, Z.; Han, Y.; Han, X.; Zhang, K.; Chang, Y.; Hu, Z.; Qi, H. Journal of the Neurological Sciences Upstream regulators and downstream effectors of NF-κ B in Alzheimer ’ s disease. J. Neurol. Sci. 2016, 366, 127–134.
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167.
- Shuaib, A.; Hartwell, A.; Kiss-Toth, E.; Holcombe, M. Multi-compartmentalisation in the MAPK signalling pathway contributes to the emergence of oscillatory behaviour and to ultrasensitivity. PLoS ONE 2016, 11.
- Tian, T.; Harding, A. How MAP kinase modules function as robust, yet adaptable, circuits. Cell Cycle 2014, 13, 2379–2390.
- Mishra, P.; Günther, S. New insights into the structural dynamics of the kinase JNK3. Sci. Rep. 2018, 8, 9435.
- Rudalska, R.; Dauch, D.; Longerich, T.; McJunkin, K.; Wuestefeld, T.; Kang, T.-W.; Hohmeyer, A.; Pesic, M.; Leibold, J.; von Thun, A.; et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat. Med. 2014, 20, 1138–1146.
- Coffey, E.T. Nuclear and cytosolic JNK signalling in neurons. Nat. Rev. Neurosci. 2014, 15, 285–299.
- Yue, J.; López, J.M. Understanding MAPK signaling pathways in apoptosis. Int. J. Mol. Sci. 2020, 21, 2346.
- Davis, R.J. Signal Transduction by the JNK Group of MAP Kinases. Cell 2000, 103, 239–252.
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257.
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776.
- Iqbal, K.; Zaidi, T.; Wen, G.Y.; Grundke-Iqbal, I.; Merz, P.A.; Shaikh, S.S.; Wisniewski, H.M.; Alafuzoff, I.; Winblad, B. Defective brain microtubule assembly in alzheimer’s disease. Lancet 1986, 328, 421–426.
- Li, H.; Zhu, H.; Xu, C.J.; Yuan, J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998, 94, 491–501.
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80.
- Schroeter, H.; Boyd, C.S.; Ahmed, R.; Spencer, J.P.E.; Duncan, R.F.; Rice-Evans, C.; Cadenas, E. c-Jun N-terminal kinase (JNK)-mediated modulation of brain mitochondria function: New target proteins for JNK signalling in mitochondrion-dependent apoptosis. Biochem. J. 2003, 372, 359–369.
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541.