| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Aigerim Soltabayeva | + 1505 word(s) | 1505 | 2021-02-02 09:55:07 | | | |
| 2 | Karina Chen | -27 word(s) | 1478 | 2021-02-05 04:32:09 | | |
Video Upload Options
Plant growth and development is adversely affected by different kind of stresses. One of the major abiotic stresses, salinity, causes complex changes in plants by influencing the interactions of genes.
1. Introduction
Growth and development of plants are affected by various stresses. Salinity is one of the major abiotic stress which adversely affects the overall growth and yield of crops [1][2][3]. It is estimated that >1 billion ha of the world land is salinized [4] and continued salinization of the ever-decreasing agricultural land further exacerbates food insecurity as human population surges. Some of the major world crops such as maize, wheat, rice, tomato and sunflower are reviewed here where, salinity resulted in the reduction of the yield [5][6][7][8]. The compromised performance causing poor yield could be due to the reduction in photosynthesis efficiency, chlorophyll, total protein, biomass, stomata closure and increasing the oxidative stress [9].
To improve productivity in salt-affected soils, selection and adoption of plant varieties with high salt tolerance has always been a preferred choice [10][11]. This selection is based on morphological, physiological and molecular markers. Among morphological markers, root or shoot morphology, visible early senescence, biomass of grains is some of the important parameters that are considered. Physiological and biochemical markers examine chlorophyll content, accumulation of proline, sucrose, stress protectants, membrane stability and hormones content [12]. These physiological markers, especially hormonal, polyamine and proline changes in plants are important to increase salt tolerance of plants. For example, such can be boosted by exogenous treatments with hormones, glycine betaine, proline, polyamines, paclobutrazol, nanoparticles [13]. The molecular markers include salt stress tolerant genes, transcription factors, metabolic pathway related genes [9][12]. These molecular markers have led to significant progress in genetic engineering of plants with salt tolerance [9]. Altogether, all stress markers in plants help in identification of specific genes involved in salt tolerance [9].
2. Evaluation of Salinity Stress in Plants by Different Stress Markers
Depending on the concentration and duration, generally, salinity affects all the plants, some of which, like Arabidopsis and tomato are more sensitive, whereas others such as wheat, rice, rye grass and so forth are less sensitive. Nevertheless, the changes at molecular, physiological, morphological level under salinity stress have similar trends (either increase or decrease) for the discussed crop plants. The measurements of Na+ and K+ ions content in plants give strong proof for salinity stress. Other stress signs may also provide the information related to salinity strength and time of exposure. For example, the morphological stress markers such as relative weight changes and germination may predict the moderate and toxic level of salinity, respectively . Monitoring the morphological changes coupled with Machine learning approaches could prevent salt stress in plants in smart greenhouse. In addition, evaluating salt sensitive (Tomato or Arabidopsis) and tolerant (salicornia) plants side-by-side in a smart greenhouse could reliably predict the ability of the examined plant to tolerate the extent of salt stress. For example, if the plant being studied suffers similar to tomato or Arabidopsis while salicornia growth remains unaffected, the salinity level in the soil is likely to in the range of 100–300 mM NaCl.
The physiological traits such as chlorophyll content, RWC, electrolyte leakage, stomatal conductance, water potential, proline, glycine betaine change on application of 100 mM NaCl after several days. The molecular markers manifest the 100 mM salt stress level prior to physiological changes. These can be detected in minutes or longer time. The ROS changes and ROS related enzyme changes are also early recognizable markers for stress [14][15][16][17][18][19][20][21]. Generally, the ROS level changes have wave shape changes during the time of stress exposure, this fluctuation of ROS occurs in stressed and also in normal conditions [22][23]. Thus, to define the fluctuation in ROS level under moderate stress, it is better to measure ROS level in time-series rather than image-type measurements, which is a snapshot measurement only for one time point [24]. Spectrometric or staining methods are commonly used for ROS detection under salinity stress [25] but also the imaging system are rapidly developing for monitoring redox state of the plant cell [26][27][28] allowing measurement of ROS changes in vivo [29].
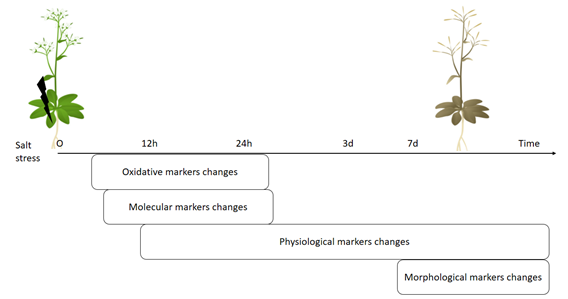
Basically, these molecular, physiological and morphological changes in plants follow the order, where the first changes by stress will be visualized by molecular, followed by biochemical then physiological and at last by morphological markers (Figure 1). Additionally, the ROS molecule plays an important role in signaling for stress and thus, these oxidative stress markers changes are detectable at similar time with molecular markers changes after exposure by salt application. Therefore, each stress marker has order in terms of time observation after stress, where the oxidative and molecular stress markers are early sensors for stress compared with other markers but they will not specify stress type.
Figure 1. Scheme of sequence of changes at different levels in plants triggered by salt stress.
3. Prediction and Identification of Stressed Plants Using Deep Learning Approaches
Machine learning techniques are developing rapidly for agricultural needs such as for plant recognition, plant or fruits counting, classification of crop types, phenotyping of various plant species, classification of mutants, leaf counting, identification of vein patterns and leaf characteristics detection of plant diseases, weed control, as well as the prediction of biotic stresses in plant leaves [30]. Basically, these approaches analyze big data of images, from monitoring various morphological changes in plants to identifying and/or classifying and/or phenotyping plants [30][31]. There are so many different machine learning approaches out there but some are frequently applied in plants, such as Artificial Neural Networks (ANN), Logistic Regression, Random Forest, Support Vector Machines (SVM), K-Nearest Neighbors (KNN) and Deep Learning are commonly used in prediction and/or classification and/or detection of stress in plants [30][32][33]. Among these different machine learning approaches, the deep learning models such as Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), Long Short-Term Memory (LSTM) are recently on the increase for use in imaging analysis. This is because the CNN has shown great accuracy in finding specific patterns in image data, so it is mainly used for identification and classification of different damages in plant leaves, especially for searching the damages caused by biotic and abiotic stimuli [30]. The other models, RNN and LSTM, are also valuable in the analysis of time series image data [30], which is important for prediction of damages in plant leaves. It has also been pointed out that various combinations of deep learning approaches can be used for classification and prediction of plant characteristics [34] and these combinations of different models can be used in the future for accurate diagnosis of signs of stress in plants for smart greenhouse procedures.
Generally, all these high-throughput phenotyping technologies are based on the analysis of different type of images such as RGB imaging, near-infrared imaging, infrared thermal imaging and fluorescence imaging [35][36][37]. These prediction of plant diseases and pest attacks or environmental impact on plants by machine learning approaches are mostly focused on identification of visual symptoms of biotic damage [38], which are discussed as morphological stress markers. As mentioned above, these morphological signs appear in plants later than physiological, oxidative and molecular stress markers (Figure 1). Currently, deep learning approaches are beginning to combine morphological stress markers data (visible signs in leaves) with physiological stress markers such as transpiration rate, biomass, water content, biochemical components (sodium concentration), photosynthetic efficiency, caratenoides [39][40][41]. However, to the best of our knowledge, there is still no extensive research on deep learning approaches for predicting abiotic or biotic stress in plants that use a combination of oxidative, molecular and morphological markers. In addition, using deep learning analysis for attached or detached leaves and whether these leaves are mature or young for better prediction has not been done yet. As previously mentioned, physiological and morphological stress marker analysis of older leaves (lower leaves) shows greater changes under salinity stress rather than young leaves but it has not yet been shown how this could affect prediction or identification damage using deep learning approaches. In addition, deep learning approaches for predicting salinity stress in plants have only been applied for a single plant species (Barley or Spinach or Okra) [42][43][44][45]. It would be interesting if machine learning approaches were applied in different plant species, specifically to the salt sensitive (Tomato or Arabidopsis) and tolerant (salicornia) plants, for their identification and prediction of salinity stress in plant . Additionally, it successfully generated different transgenic plants with different gene modifications, for example, it generated fused constructions of the gene promoter region with GFP proteins [46]. These transgenic plants with GFP protein, could be useful for evaluation of molecular stress markers and prediction of salinity stress in plants. Thus, we believe that the following suggestions have potential for application through deep learning approaches for stress prediction in plants: a) analysis of images data with other stress markers such as oxidative and/or molecular markers, b) emphasis on analysis of mature leaves versus young leaves, c) use of control plant data such as stress-sensitive or stress-tolerant plants or the transgenic plant promoter fused with GFP and other fluorescent markers.
References
- Flowers, T.J.; Yeo, A.R. Effects of salinity on plant growth and crops yields. In Environmental Stress in Plants; Cherry, J.H., Ed.; NATO ASI Series; Springer: Berlin, Germany, 1989; Volume G19, pp. 101–119.
- Ali, Y.; Aslam, Z.; Ashraf, M.Y.; Tahir, G.R. Effect of salinity on chlorophyll concentration, leaf area, yield and yield components of rice genotypes grown under saline environment. Int. J. Environ. Sci. Technol. 2004.
- Yermiyahu, U.; Ben-Gal, A.; Keren, R.; Reid, R.J. Combined effect of salinity and excess boron on plant growth and yield. Plant Soil 2008.
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019.
- Khatoon, A.; Qureshi, M.S.; Hussain, M.K. Effect of Salinity on Some Yield Parameters of Sunflower (Helianthus annuus L.). Int. J. Agric. Biol. 2000, 2, 382–384.
- Abd El-Kader, A.A.; Mohamedin, A.A.M.; Ahmed, M.K.A. Growth and Yield of Sunflower as Affected by Different Salt Affected Soils. Int. J. Agric. Biol. 2006, 8, 583–587.
- Chamekh, Z.; Ayadi, S.; Karmous, C.; Trifa, Y.; Amara, H.; Boudabbous, K.; Yousfi, S.; Serret, M.D.; Araus, J.L. Comparative effect of salinity on growth, grain yield, water use efficiency, δ13C and δ15N of landraces and improved durum wheat varieties. Plant Sci. 2016.
- Elkhatib, H.A.; Elkhatib, E.A.; Khalaf Allah, A.M.; El-Sharkawy, A.M. Yield Response of Salt-Stressed Potato to Potassium Fertilization: A Preliminary Mathematical Model. J. Plant Nutr. 2004.
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics 2014.
- Ondrasek, G.; Rengel, Z.; Veres, S. Soil Salinisation and Salt Stress in Crop Production. In Abiotic Stress in Plants—Mechanisms and Adaptations; Shanker, A.K., Venkateswarlu, B., Eds.; InTech: Rijeka, Croatia, 2011; pp. 171–190. ISBN 978-953-307-394-1.
- Sytar, O.; Brestic, M.; Zivcak, M.; Olsovska, K.; Kovar, M.; Shao, H.; He, X. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. Sci. Total Environ. 2017.
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681.
- Mbarki, S.; Sytar, O.; Cerda, A.; Zivcak, M.; Rastogi, A.; He, X.; Zoghlami, A.; Abdelly, C.; Brestic, M. Strategies to mitigate the salt stress effects on photosynthetic apparatus and productivity of crop plants. In Salinity Responses and Tolerance in Plants, Volume 1: Targeting Sensory, Transport and Signaling Mechanisms; Springer International Publishing: Cham, Switzerland, 2018; pp. 85–136. ISBN 9783319756714.
- Ellouzi, H.; Ben Hamed, K.; Cela, J.; Munné-Bosch, S.; Abdelly, C. Early effects of salt stress on the physiological and oxidative status of Cakile maritima (halophyte) and Arabidopsis thaliana (glycophyte). Physiol. Plant. 2011.
- Guan, Q.; Takano, T.; Liu, S. Genetic transformation and analysis of rice OsAPx2 gene in Medicago sativa. PLoS ONE 2012.
- Poór, P.; Kovács, J.; Borbély, P.; Takács, Z.; Szepesi, Á.; Tari, I. Salt stress-induced production of reactive oxygen- and nitrogen species and cell death in the ethylene receptor mutant Never ripe and wild type tomato roots. Plant Physiol. Biochem. 2015.
- Poór, P.; Kovács, J.; Szopkó, D.; Tari, I. Ethylene signaling in salt stress- and salicylic acid-induced programmed cell death in tomato suspension cells. Protoplasma 2013.
- Kataria, S.; Baghel, L.; Guruprasad, K.N. Pre-treatment of seeds with static magnetic field improves germination and early growth characteristics under salt stress in maize and soybean. Biocatal. Agric. Biotechnol. 2017.
- Hanqing, F.; Yifeng, W.; Hongyu, L.; Rongfang, W.; Kun, S.; Lingyun, J. Salt stress-induced expression of rice AOX1a is mediated through an accumulation of hydrogen peroxide. Biologia (Bratisl) 2010.
- Li, J.; Jia, H.; Wang, J.; Cao, Q.; Wen, Z. Hydrogen sulfide is involved in maintaining ion homeostasis via regulating plasma membrane Na+/H+ antiporter system in the hydrogen peroxide-dependent manner in salt-stress Arabidopsis thaliana root. Protoplasma 2014.
- Liu, S.G.; Zhu, D.Z.; Chen, G.H.; Gao, X.Q.; Zhang, X.S. Disrupted actin dynamics trigger an increment in the reactive oxygen species levels in the Arabidopsis root under salt stress. Plant Cell Rep. 2012.
- Bailey-Serres, J.; Mittler, R. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006.
- Vestergaard, C.L.; Flyvbjerg, H.; Møller, I.M. Intracellular signaling by diffusion: Can waves of hydrogen peroxide transmit intracellular information in plant cells? Front. Plant Sci. 2012.
- Ortega-Villasante, C.; Burén, S.; Blázquez-Castro, A.; Barón-Sola, Á.; Hernández, L.E. Fluorescent in vivo imaging of reactive oxygen species and redox potential in plants. Free Radic. Biol. Med. 2018, 122, 202–220.
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010.
- Jiang, K.; Schwarzer, C.; Lally, E.; Zhang, S.; Ruzin, S.; Machen, T.; Remington, S.J.; Feldman, L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 2006.
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007.
- Morgan, M.J.; Lehmann, M.; Schwarzländer, M.; Baxter, C.J.; Sienkiewicz-Porzucek, A.; Williams, T.C.R.; Schauer, N.; Fernie, A.R.; Fricker, M.D.; Ratcliffe, R.G.; et al. Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol. 2008.
- Navarro, J.M.; Martínez, V.; Carvajal, M. Ammonium, bicarbonate and calcium effects on tomato plants grown under saline conditions. Plant Sci. 2000.
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90.
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep Learning for Plant Stress Phenotyping: Trends and Future Perspectives. Trends Plant Sci. 2018, 23, 883–898.
- Douarre, C.; Schielein, R.; Frindel, C.; Gerth, S.; Rousseau, D. Deep learning based root-soil segmentation from X-ray tomography. bioRxiv 2016.
- Akar, Ö.; Güngör, O. Classification of multispectral images using Random Forest algorithm. J. Geod. Geoinf. 2012.
- Ho Tong Minh, D.; Ienco, D.; Gaetano, R.; Lalande, N.; Ndikumana, E.; Osman, F.; Maurel, P. Deep Recurrent Neural Networks for Winter Vegetation Quality Mapping via Multitemporal SAR Sentinel-1. IEEE Geosci. Remote Sens. Lett. 2018.
- Feng, X.; Yu, C.; Chen, Y.; Peng, J.; Ye, L.; Shen, T.; Wen, H.; He, Y. Non-destructive determination of shikimic acid concentration in transgenic maize exhibiting glyphosate tolerance using chlorophyll fluorescence and hyperspectral imaging. Front. Plant Sci. 2018.
- Biju, S.; Fuentes, S.; Gupta, D. The use of infrared thermal imaging as a non-destructive screening tool for identifying drought-tolerant lentil genotypes. Plant Physiol. Biochem. 2018.
- Chen, D.; Shi, R.; Pape, J.M.; Neumann, K.; Arend, D.; Graner, A.; Chen, M.; Klukas, C. Predicting plant biomass accumulation from image-derived parameters. Gigascience 2018.
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90.
- Alhnaity, B.; Pearson, S.; Leontidis, G.; Kollias, S. Using deep learning to predict plant growth and yield in greenhouse environments. arXiv 2019, arXiv:1907.00624.
- Altangerel, N.; Ariunbold, G.O.; Gorman, C.; Alkahtani, M.H.; Borrego, E.J.; Bohlmeyer, D.; Hemmer, P.; Kolomiets, M.V.; Yuan, J.S.; Scully, M.O. In vivo diagnostics of early abiotic plant stress response via Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2017.
- Gao, G.; Tester, M.A.; Julkowska, M.M. The Use of High-Throughput Phenotyping for Assessment of Heat Stress-Induced Changes in Arabidopsis. Plant Phenomics 2020.
- Feng, X.; Zhan, Y.; Wang, Q.; Yang, X.; Yu, C.; Wang, H.; Tang, Z.Y.; Jiang, D.; Peng, C.; He, Y. Hyperspectral imaging combined with machine learning as a tool to obtain high-throughput plant salt-stress phenotyping. Plant J. 2020.
- Behmann, J.; Steinrücken, J.; Plümer, L. Detection of early plant stress responses in hyperspectral images. ISPRS J. Photogramm. Remote Sens. 2014.
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Klukas, C. Dissecting the phenotypic components of crop plant growthand drought responses based on high-throughput image analysis w open. Plant Cell 2014.
- Kersting, K.; Xu, Z.; Wahabzada, M.; Bauckhage, C.; Thurau, C.; Roemer, C.; Ballvora, A.; Rascher, U.; Leon, J.; Pluemer, L. Pre-symptomatic prediction of plant drought stress using Dirichlet-aggregation regression on hyperspectral images. In Proceedings of the Twenty-Sixth AAAI Conference on Artificial Intelligence, Toronto, ON, Canada, 22–26 July 2012; AAAI Publications: Toronto, ON, Canada, 2012; pp. 302–308.
- Chiu, W.L.; Niwa, Y.; Zeng, W.; Hirano, T.; Kobayashi, H.; Sheen, J. Engineered GFP as a vital reporter in plants. Curr. Biol. 1996.