
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Christopher McMillan | + 2537 word(s) | 2537 | 2021-01-26 07:35:28 | | | |
| 2 | Catherine Yang | Meta information modification | 2537 | 2021-02-02 04:32:21 | | |
Video Upload Options
Influenza viruses are Orthomyxovirus species belonging to the Orthomyxoviridae family. While influenza viruses can be classified into four genera (A, B, C and D), only influenza A and B viruses cause clinical disease in humans.
1. Introduction
Influenza viruses are characterized based on their surface proteins—hemagglutinin (HA) and neuraminidase (NA). There are currently 18 known serotypes/antigenic types of HA (Figure 1a) and 11 known of NA. Almost all human influenza infections are caused by H1- and H3-containing strains (H1N1 and H3N2), with two lineages of influenza B virus—Victoria and Yamagata—also circulating globally on a seasonal basis [1]. Of particular concern, however, are zoonotic events in which highly pathogenic avian influenza viruses such as H5N1, H7N9 and H9N2 viruses are transmitted from avian species to humans. Although limited human-to-human transmission is observed, case fatality rates are significantly higher than seasonal infections (e.g., H5N1, ~53%; H7N9, ~32%) [2]. Furthermore, evidence suggests only minor genetic changes are required to allow increased replication rates in human cells, which could lead to more efficient human-to-human transmission and a subsequent pandemic [3][4]. Additionally, emergence of reassortant viruses whereby entire genome segments are reassorted between viruses upon co-infection of a host with different strains is another mechanism by which pandemic influenza viruses can emerge. Such an event was responsible for the H1N1 pandemic of 2009 and has seen H5N6 viruses emerge in aquatic duck populations more recently [5].
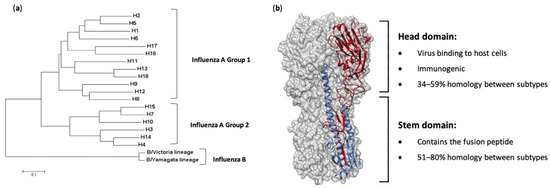
Figure 1. Phylogeny and structure of influenza virus hemagglutinin. (a) A rooted phylogenetic tree based on the amino acid sequences of hemagglutinin (HA) sequences from influenza A and B viruses, adapted from Noh et al. [6]. (b) The structure of the ectodomain of the influenza hemagglutinin protein from the A/California/04/2009(H1N1) virus (PDB ID 3LZG). One HA monomer is shown with the HA1 subunit in red and the HA2 subunit in blue.
To control the impacts of influenza virus infections, vaccination is the best possible intervention. The influenza HA protein (Figure 1b) has been the primary target for vaccines, as it is large, readily accessible on the virus surface, is essential for virus binding and infection of host cells and is the major target of neutralizing antibodies [7]. While antibodies are produced against other influenza proteins during virus infection, and some of these antigens have been trialed previously as vaccine candidates, anti-HA antibodies are the most abundant and protective [8][9][10][11][12]. As such, current licensed influenza vaccines aim to induce HA-specific antibodies. Traditional influenza vaccines, of which approximately 500–800 million doses are produced annually [13], are created by inactivation and splitting of influenza viruses that have been propagated in hens’ embryonated eggs. Each vaccine dose is measured by HA content, and a standard adult dose contains from 15 to 60 µg of each HA (i.e., H1, H3 and either one or the two HAs from the two influenza B lineages, depending on whether it is a tri- or quadrivalent vaccine) [14]. In people over 65 years of age, high-dose vaccines containing 60 µg of HA from each strain, or suitably adjuvanted influenza vaccines, are recommended [15]. The virion surface-displayed HA molecule is a virion membrane-anchored trimer that comprises two structural elements: a distal, highly variable head domain that contains the receptor binding site and a membrane-proximal domain that shows a high degree of homology between strains and is referred to as the stem (Figure 1b).
Unfortunately, inactivated vaccines have only proven to be effective against homologous virus strains, owing to the apparent immunodominance of the highly variable HA head domain rather than the more conserved stem domain, with the majority of antibodies induced being directed towards this region [16][17][18][19][20]. Indeed, the HA head domain evolves at a faster rate than the stem domain [21], leading to a constant arms race to update and re-administer vaccines annually in order to keep up with this virus evolution. Additionally, such vaccines would likely offer little to no protection in the event of a zoonotic spillover event, meaning a new vaccine would have to be manufactured at a rapid speed. Ideally, a universal influenza vaccine capable of providing long-lasting protection against both seasonal infections as well as potential pandemic viruses should be available. If such an ambitious goal were to be achieved, new approaches to influenza virus vaccine development are required.
2. Hemagglutinin Stem-Based Vaccines
Due to the immunodominance of the head domain, attempts to elicit a broadly protective immune response have focused on the more highly conserved HA stem domain [21]. Many monoclonal antibodies directed towards the HA stem domain have proven to be broadly protective, either within one of the phylogenetic groups outlined in Figure 1a or even providing universal protection from both group 1 and group 2 viruses [22][23][24][25][26][27][28][29]. This protection is mediated either through Fc effector functions or inhibition of the low pH-induced conformational changes necessary for membrane fusion.
To create these HA stem vaccine candidates, many approaches have been trialed. Initial studies designed HA stem constructs and delivered these as DNA or virus-like particle (VLP) vaccines [30]. Vaccination of mice with these VLPs was able to protect from homologous virus challenge and induced cross-reactive antibody responses capable of binding to heterologous HAs [30].
Subsequent studies utilized recombinant protein technology, incorporating trimerization domains (foldon or GCN4) as well as rational stabilizing mutations to produce soluble recombinant HA (rHA) stem vaccine candidates [30][31][32][33][34][35][36][37][38][39]. One such HA stem vaccine termed mini-HA #4900, which was based on H1 HA, was shown to provide protection against heterologous H1N1 and H5N1 challenge in a mouse model with no clinical symptoms or weight loss observed. Furthermore, sera from immunized mice contained antibodies able to bind to H1, H3, H5, H7 and H9 HAs, with neutralizing activity against an H5N1 virus also observed. These antibodies were also able to mediate antibody-dependent cellular cytotoxicity (ADCC), a key mechanism for broadly protective antibodies against influenza viruses. Upon challenge in cynomolgus monkeys, a significant reduction in fever was observed in vaccinated animals compared to control animals; however, no difference was observed in tracheal viral loads. This illustrates that well-designed HA stem constructs can induce broadly cross-reactive immune responses, though they do not necessarily reduce viral replication.
Stem-only HA constructs have also been designed from H3 HAs. Mallajosyula and colleagues [35] used sequence conservation to guide the design of multiple HA stem constructs and included the GCN4 isoleucine zipper or bacteriophage T4 foldon trimerization domain to enhance folding. Vaccination of mice with these constructs induced antibodies capable of binding to multiple H1, H3 and H7 HAs, and neutralizing a heterologous H3N2 pseudovirus. When assessed in an in vivo mouse model, however, only partial (40–50%) protection was observed following challenge with a homologous H3N2 virus, highlighting the challenges of achieving protection with stem-only HA constructs.
While most stem-only constructs provide protection from divergent strains, this was seen mostly within the same phylogenetic group. Inter-group protection has been achieved, however, with HA stem proteins based on H1 or H5 HAs [37]. In this study, vaccination with H1 or H5 vaccines provided protection from a H3N2 virus challenge in mice, with 40% and 80% survival after vaccination with H1 or H5 stem vaccines reported, respectively. While survival was evident after this H3N2 challenge, the viral load in the lungs was not reduced by vaccination, consistent with previous studies involving HA stem constructs.
To improve on the modest protection observed by many groups with headless rHAs, attempts to model pre-existing memory immune responses to the stem domain have been made. It was hypothesized that as some human populations have low levels of stem-reactive antibodies, immunization with stem-only rHA could selectively boost this antibody population, leading to broader protection [14][15][39]. To establish an animal model system which accounted for this memory, Wohlbold et al. immunized mice with a DNA vaccine encoding a chimeric HA consisting of the head domain from H9 HA and the stem domain from H1 HA. Such a vaccination strategy has been shown to induce a weak anti-H1 stem response [38]. When mice were primed with the DNA vaccine before immunization with stem-only rHA, complete protection was observed from homologous H1N1 challenge compared to just 40% from immunization with stem-only rHA alone. In a heterologous H6N1 virus challenge, 100% protection was seen when primed with the DNA vaccine, and 60% protection upon H5N1 virus challenge [38].
A similar approach was used by Ibanez et al. [40], where a stem-only HA vaccine based on an equine H3N8 influenza virus was utilized in DNA and subunit vaccine forms. Mice were vaccinated with a stem-only DNA vaccine (encoding the stem-only HA with a GCN4 trimerization motif) or a subunit protein vaccination boost (containing prokaryote-expressed HA stem with no trimerization domain), or prime-boost combinations of the two [40]. Using this strategy, both the DNA vaccine and the subunit vaccine, as well as DNA prime followed by the subunit boost, and vice versa, yielded 100% protection from homologous virus challenge. When a homosubtypic human H3N2 virus was used, only the regime using a subunit prime followed by a DNA vaccine boost showed 100% protection, with all other regimes showing only partial (20–80%) protection [40].
These studies suggest that a small amount of pre-existing anti-stem immunity, as is likely present in some of the human population, improves the effectiveness of stem-only influenza vaccines and provides a more cross-reactive antibody response. These results may provide a framework for the future application of headless rHAs as potential vaccine candidates in humans. Indeed, clinical trials assessing a stem-only vaccine candidate are currently underway (NCT03814720), which will be greatly beneficial to assess the impact of such vaccines in the human population.
3. Chimeric Hemagglutinin Vaccine Candidates
Another approach to inducing stem-specific antibodies involves the use of chimeric HAs (cHAs). These cHAs contain a stem domain from one subtype (e.g., H1 or H3) and a head domain from another, foreign subtype to which the subject is naïve (e.g., H5, H6 or H9) [41]. It was hypothesized that by sequential vaccination with cHAs with a common stem domain but different head domains, stem-specific antibodies would be selectively boosted and, thus, broader protection would result. This is outlined in Figure 2.
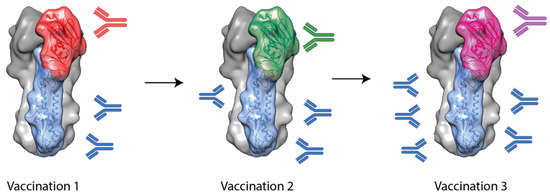
Figure 2. A schematic outlining chimeric HA-based vaccination approaches. A subject is immunized with a chimeric HA with an exotic head domain but a stem domain for the target virus. Following from this, a second vaccination can be administered with a chimeric HA with a consistent stem domain but another, different, exotic head domain. This can continue in order to selectively boost antibody responses towards the conserved stem domain while not boosting head-specific antibodies.
Studies using foldon-stabilized recombinant cHAs have shown that this technique has promise [42][43]. In these studies, mice were vaccinated three times with cHAs with common stem domains (e.g., H1 or H3 stem) but foreign head domains (e.g., H9, H6 or H5). Upon challenge with heterosubtypic viruses, from within the same HA phylogenetic group, cHA vaccination regimes were able to provide complete protection, though they failed to provide protection from intergroup challenge viruses [42][43]. This suggests the stem-specific antibodies induced by cHAs are mostly restricted to one hemagglutinin group.
While most efforts towards a universal influenza vaccine have focused on influenza A viruses, this cHA technology has been applied to influenza B viruses [44]. These candidate vaccines, which contained head domains from influenza A viruses with stem domains from influenza B viruses, could protect from a lethal challenge with diverse influenza B viruses from both Victoria and Yamagata lineages [44]. Analysis of the serum indicated that this protection was largely due to antibody effector functions such as ADCC rather than virus neutralization. This observation is consistent with data from studies on stem-specific monoclonal antibodies (mAbs), where their main in vivo mechanism of action was via antibody effector functions [22][45].
HA chimeras have been made as part of whole viruses as well as soluble recombinant HAs and, thus, can be used as split virus vaccines, live-attenuated influenza virus vaccines or subunit vaccines [41][42][46]. One such study utilized this cHA technology in a split virus vaccine modality [46]. Mice were first primed with a monovalent inactivated H1N1pdm09 vaccine before vaccination with H1 chimeric rHAs containing either a H5 or H8 head domain [46]. This approach induced higher stem-specific antibody levels when compared to current seasonal vaccines containing regular HA. These stem-specific antibodies were able to bind to H2 and H18 HAs in vitro [46]. The serum from immunized mice was able to provide complete protection from a heterologous virus challenge in a passive transfer experiment, where acceptor mice received sera from donor mice that were vaccinated with the chimeric regime outlined earlier before a challenge with a chimeric virus (containing a H1 HA chimera with an H9 head domain and an N3 NA protein—to ensure the only antibodies present from the donor mice were towards the conserved H1 stem domain) [46]. This study demonstrates that the cHA technology provides some level of stem-specific immunity that can protect from challenge with heterologous HA viruses.
Building on these pre-clinical data, data from a Phase I clinical trial were also recently published using this approach in the form of a prime/boost strategy with live attenuated and inactivated cHA virus vaccines [47]. Here, it was found that immunization with AS03 as an adjuvant elicited broadly cross-reactive antibodies directed towards the stem domain of HA. This important finding highlights the utility of the cHA approach in the human population.
With a similar objective as the cHA strategy, other approaches have focused on replacing more distinct antigenic sites in the HA head region rather than the entire domain [48][49]. By substituting immunodominant major antigenic sites of the H3 protein with corresponding sequences from exotic HAs, “mosaic” HAs are created. These HAs were then incorporated into reassortant viruses and subsequently inactivated for use as vaccines, where they induced broadly reactive stem-specific antibodies as well as head-specific neutralizing antibodies. Additionally, protection was afforded from challenge with historical H3N2 virus strains [48]. This approach has also been validated with influenza B viruses as a subunit vaccine approach, where cross-protection against heterologous influenza B virus strains was observed in mice [49].
4. Conclusions and Outlook
The next generation of influenza vaccines would ideally be a universal solution—capable of providing complete protection from all influenza A and B viruses. While this is a lofty goal that is perhaps currently out of reach, research has been focused on vaccines that may lead us at least somewhat closer to this target. Such a vaccine would provide improved protection from drifted seasonal viruses that do not exactly match the vaccine strains, thereby limiting the impact of seasonal influenza virus infections while also providing some degree of cross-protection to novel, heterologous strains such as highly pathogenic avian influenza (HPAI) viruses that might cause a pandemic. This vaccine would likely need to target other viral antigens apart from HA. A novel vaccine containing multiple influenza virus antigens in combination, such as HA, NA and M2e, could be more likely to achieve the goal of improved protection from drifted and heterologous strains. This vaccine could also provide partial protection to slow the spread and impact of any potential pandemic strain that emerges, buying valuable time until a more potent strain-specific vaccine is made and is readily available. In such a case, rapid platform technologies capable of responding quickly upon virus discovery, such as mRNA vaccines, would prove valuable in the rapid production of a vaccine to slow the pandemic.
References
- World Health Organisation. Influenza (Seasonal). Available online: http://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 28 July 2020).
- World Health Organisation. Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2020. Available online: https://www.who.int/influenza/human_animal_interface/2020_01_20_tableH5N1.pdf?ua=1 (accessed on 17 March 2020).
- Manz, B.; de Graaf, M.; Mogling, R.; Richard, M.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Fouchier, R.A. Multiple Natural Substitutions in Avian Influenza A Virus PB2 Facilitate Efficient Replication in Human Cells. J. Virol. 2016, 90, 5928–5938.
- Yamaji, R.; Yamada, S.; Le, M.Q.; Li, C.; Chen, H.; Qurnianingsih, E.; Nidom, C.A.; Ito, M.; Sakai-Tagawa, Y.; Kawaoka, Y. Identification of PB2 mutations responsible for the efficient replication of H5N1 influenza viruses in human lung epithelial cells. J. Virol. 2015, 89, 3947–3956.
- Wong, F.Y.; Phommachanh, P.; Kalpravidh, W.; Chanthavisouk, C.; Gilbert, J.; Bingham, J.; Davies, K.R.; Cooke, J.; Eagles, D.; Phiphakhavong, S.; et al. Reassortant highly pathogenic influenza A(H5N6) virus in Laos. Emerg. Infect. Dis. 2015, 21, 511–516.
- Noh, J.Y.; Kim, W.J. Influenza vaccines: Unmet needs and recent developments. Infect. Chemother. 2013, 45, 375–386.
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 1981, 289, 366–373.
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K.; et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214.
- Sui, Z.; Chen, Z.; Chen, Q.; Fang, F.; Zheng, M. Cross-protection against influenza virus infection by intranasal administration of M1-based vaccine with chitosan as an adjuvant. Vaccine 2010, 28, 7690–7698.
- Park, M.-H.; Chang, J. Immunogenicity and protective efficacy of a dual subunit vaccine against respiratory syncytial virus and influenza virus. Immune Netw. 2012, 12, 261.
- Doyle, T.M.; Hashem, A.M.; Li, C.; Van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013, 100, 567–574.
- Uddback, I.E.; Steffensen, M.A.; Pedersen, S.R.; Nazerai, L.; Thomsen, A.R.; Christensen, J.P. PB1 as a potential target for increasing the breadth of T-cell mediated immunity to Influenza A. Sci. Rep. 2016, 6, 35033.
- Partridge, J.; Kieny, M.P. Global production capacity of seasonal influenza vaccine in 2011. Vaccine 2013, 31, 728–731.
- U.S. Food and Drug Administration. Influenza Virus Vaccine for the 2014–2015 Season. Available online: http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Post-MarketActivities/LotReleases/ucm397090.htm (accessed on 2 March 2015).
- Centres for Disease Control and Prevention. Flu & People 65 Years and Older. Available online: https://www.cdc.gov/flu/highrisk/65over.htm (accessed on 3 December 2020).
- Ellebedy, A.H.; Krammer, F.; Li, G.M.; Miller, M.S.; Chiu, C.; Wrammert, J.; Chang, C.Y.; Davis, C.W.; McCausland, M.; Elbein, R.; et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 13133–13138.
- Nachbagauer, R.; Wohlbold, T.J.; Hirsh, A.; Hai, R.; Sjursen, H.; Palese, P.; Cox, R.J.; Krammer, F. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J. Virol. 2014, 88, 13260–13268.
- Corti, D.; Suguitan, A.L., Jr.; Pinna, D.; Silacci, C.; Fernandez-Rodriguez, B.M.; Vanzetta, F.; Santos, C.; Luke, C.J.; Torres-Velez, F.J.; Temperton, N.J.; et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Investig. 2010, 120, 1663–1673.
- Thomson, C.A.; Wang, Y.; Jackson, L.M.; Olson, M.; Wang, W.; Liavonchanka, A.; Keleta, L.; Silva, V.; Diederich, S.; Jones, R.B.; et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Front. Immunol. 2012, 3, 87.
- Margine, I.; Hai, R.; Albrecht, R.A.; Obermoser, G.; Harrod, A.C.; Banchereau, J.; Palucka, K.; Garcia-Sastre, A.; Palese, P.; Treanor, J.J.; et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J. Virol. 2013, 87, 4728–4737.
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 10432.
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856.
- Srivastava, V.; Yang, Z.; Hung, I.F.N.; Xu, J.; Zheng, B.; Zhang, M.-Y. Identification of Dominant Antibody-Dependent Cell-Mediated Cytotoxicity Epitopes on the Hemagglutinin Antigen of Pandemic H1N1 Influenza Virus. J. Virol. 2013, 87, 5831–5840.
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151.
- Ana-Sosa-Batiz, F.; Vanderven, H.; Jegaskanda, S.; Johnston, A.; Rockman, S.; Laurie, K.; Barr, I.; Reading, P.; Lichtfuss, M.; Kent, S.J. Influenza-Specific Antibody-Dependent Phagocytosis. PLoS ONE 2016, 11, e0154461.
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610.
- Dunand, C.J.H.; Leon, P.E.; Huang, M.; Choi, A.; Chromikova, V.; Ho, I.Y.; Tan, G.S.; Cruz, J.; Hirsh, A.; Zheng, N.-Y.; et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016, 19, 800–813.
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Neu, U.; McAuliffe, J.M.; Benjamin, E.; Wachter-Rosati, L.; Palmer-Hill, F.J.; Yuan, A.Q.; Walker, P.A.; et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 2016.
- Mullarkey, C.E.; Bailey, M.J.; Golubeva, D.A.; Tan, G.S.; Nachbagauer, R.; He, W.; Novakowski, K.E.; Bowdish, D.M.; Miller, M.S.; Palese, P. Broadly Neutralizing Hemagglutinin Stalk-Specific Antibodies Induce Potent Phagocytosis of Immune Complexes by Neutrophils in an Fc-Dependent Manner. MBio 2016, 7.
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; Garcia-Sastre, A.; Palese, P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 2010, 1.
- Bommakanti, G.; Lu, X.; Citron, M.P.; Najar, T.A.; Heidecker, G.J.; ter Meulen, J.; Varadarajan, R.; Liang, X. Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J. Virol. 2012, 86, 13434–13444.
- Lu, Y.; Welsh, J.P.; Swartz, J.R. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl. Acad. Sci. USA 2014, 111, 125–130.
- Mallajosyula, V.V.; Citron, M.; Ferrara, F.; Lu, X.; Callahan, C.; Heidecker, G.J.; Sarma, S.P.; Flynn, J.A.; Temperton, N.J.; Liang, X.; et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. USA 2014, 111, E2514–E2523.
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.B.; van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306.
- Mallajosyula, V.V.A.; Citron, M.; Ferrara, F.; Temperton, N.J.; Liang, X.; Flynn, J.A.; Varadarajan, R. Hemagglutinin Sequence Conservation Guided Stem Immunogen Design from Influenza A H3 Subtype. Front. Immunol. 2015, 6, 329.
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.-J.; Kanekiyo, M.; Kong, W.-P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015.
- Valkenburg, S.A.; Mallajosyula, V.V.A.; Li, O.T.W.; Chin, A.W.H.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L.M. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016, 6, 22666.
- Wohlbold, T.J.; Nachbagauer, R.; Margine, I.; Tan, G.S.; Hirsh, A.; Krammer, F. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 2015, 33, 3314–3321.
- Wang, S.-C.; Liao, H.-Y.; Zhang, J.-Y.; Cheng, T.-J.R.; Wong, C.-H. Development of a universal influenza vaccine using hemagglutinin stem protein produced from Pichia pastoris. Virology 2019, 526, 125–137.
- Ibanez, L.I.; Caldevilla, C.A.; Paredes Rojas, Y.; Mattion, N. Genetic and subunit vaccines based on the stem domain of the equine influenza hemagglutinin provide homosubtypic protection against heterologous strains. Vaccine 2018, 36, 1592–1598.
- Hai, R.; Krammer, F.; Tan, G.S.; Pica, N.; Eggink, D.; Maamary, J.; Margine, I.; Albrecht, R.A.; Palese, P. Influenza Viruses Expressing Chimeric Hemagglutinins: Globular Head and Stalk Domains Derived from Different Subtypes. J. Virol. 2012, 86, 5774–5781.
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550.
- Krammer, F.; Margine, I.; Hai, R.; Flood, A.; Hirsh, A.; Tsvetnitsky, V.; Chen, D.; Palese, P. H3 Stalk-Based Chimeric Hemagglutinin Influenza Virus Constructs Protect Mice from H7N9 Challenge. J. Virol. 2014, 88, 2340–2343.
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91, e00286-17.
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348.
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1, 16015.
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2020.
- Broecker, F.; Liu, S.T.H.; Suntronwong, N.; Sun, W.; Bailey, M.J.; Nachbagauer, R.; Krammer, F.; Palese, P. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 2019, 4, 31.
- Sun, W.; Kirkpatrick, E.; Ermler, M.; Nachbagauer, R.; Broecker, F.; Krammer, F.; Palese, P. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol. 2019, 93, e00333-19.




