
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ruben Warkentin | + 2248 word(s) | 2248 | 2021-01-22 04:52:56 | | | |
| 2 | Lily Guo | Meta information modification | 2248 | 2021-01-28 05:31:30 | | | | |
| 3 | Lily Guo | Meta information modification | 2248 | 2021-01-28 05:31:52 | | |
Video Upload Options
Glycans—a broad term describing carbohydrates, including oligosaccharides and polysaccharides—are the third class of important biological macromolecules following nucleic acids and proteins. Glycans are found in all domains of life and in viruses. They can exist as free sugars, but are more commonly found as glycoconjugates, including proteoglycans, glycoproteins, and glycolipids. Glycans are involved in a wide variety of physiological functions and have implications in numerous infectious and non-infectious diseases, making them diagnostic and therapeutic targets. Additionally, glycans are targeted in various biotechnological and industrial applications. The broad applications of glycans have spurred interest in the development of glycan binding proteins (GBPs).
GBPs include lectins, antibodies, pseudoenzymes, and carbohydrate-binding modules (CBMs). Lectins are non-immunoglobulin proteins containing at least one non-catalytic domain that exhibits reversible carbohydrate binding. CBMs are similar to lectins, but are small binding domains typically found in lectins or carbohydrate-active enzymes (CAZymes). CAZymes can be further classified into glycoside hydrolases, glycosyltransferases, polysaccharide lyases, and carbohydrate esterases—detailed information on these enzymes is available through the Carbohydrate Active Enzymes (CAZy) database.
1. Introduction
Choosing the right protein scaffold is a crucial aspect of engineering a GBP with improved or novel binding properties. In this section we discuss examples of several protein scaffolds available for GBP engineering, including lectins, carbohydrate binding modules (CBMs), pseudoenzymes, carbohydrate-active enzymes (CAZymes), and antibody-based scaffolds (Figure 1). The definition of lectins has changed over the years[1][2], but can generally be defined as proteins that bind carbohydrates. Hence, most GBPs can be categorized as a lectin; however, for the purpose of this review, we have categorized certain GBPs separately from lectins due to their distinct characteristic folds and properties. A summary of the scaffolds discussed in this section, along with example scaffolds that have structural and binding data available, is available in Table 1.
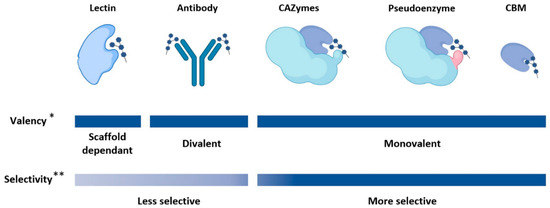
Figure 1. Valency and selectivity of protein scaffolds with glycan binding sites. The valency of lectins and antibody-based scaffold varies, as some lectins contain tandem repeat units and antibody-based scaffolds can be designed to contain only a single, or multiple variable fragments. Similarly, CBMs can be designed in tandem to increase valency. The selectivity of antibody-based scaffold is affected by the poor immunogenicity of carbohydrates. * It should be noted that the valency of lectins, antibodies, and CBMs can be altered with protein engineering. ** There are exceptions to this trend and the selectivity can be affected by valency.
2. Lectins
Lectins are carbohydrate binding proteins that are placed into sub-categories based on their folds and function: P-type, I-type, L-type, R-type, C-type, and galectins. Lectins display a wide variety of physiological functions and have biotechnological and biomedical applications—lectins have already been used in the detection and targeted treatments of human diseases such as cancer[3][4]. Here we provide a brief overview of lectins and some examples in GBP engineering. An excellent resource for detailed information on the various sub-categories of lectins can be found in the comprehensive text, Essentials of Glycobiology (specifically chapters 28 to 38)[5].
Generally, lectins have relatively low affinities for their glycan targets, with dissociation constants in the micromolar range[6][7]. This may be explained by the shallow binding interface that is observed in most lectins, causing more competitive solvent interactions. The shallow binding interface may also explain the promiscuous binding observed in lectins—glycans with similar structures often bind similar lectins. In nature, the low affinity problem is overcome by oligomerization and multivalency; in biological settings lectins tend to assemble into oligomeric structures containing multiple binding sites, allowing for higher affinities to be reached. The relatively low affinities and promiscuity of lectins in the monomeric state must be considered when selecting scaffolds for GBP engineering; however, lectins with improved binding specificity and affinity have been developed[8]. One advantage of using lectins over other protein scaffolds is that databases like UniLectin3D are available that can search for lectin scaffolds based on the glycan target[9].
3. Carbohydrate Binding Modules
Carbohydrate binding modules (CBMs)[10], also known as carbohydrate binding domains (CBDs), are non-catalytic protein domains generally found on carbohydrate-active enzymes (CAZymes). There is low sequence identity between CBMs[10], but there are conserved tertiary folds that are categorized based on their binding site topology as types A, B, or C [11]. The topologies of CBMs are characterized in type A by a planar hydrophobic surface, in type B by an extended binding cavity, and in type C by a short binding pocket—for more information on the structures of CBM types please see the extensive review by Armenta et al. [10]. For the purposes of GBP engineering, type A CBMs are suitable for binding insoluble, crystalline carbohydrates, due to the exposed planar binding interface [12]. In contrast, type B CBMs bind oligosaccharides[13], and type C CBMs bind mono and di-saccharides[14]. One attractive aspect of CBMs as GBP scaffolds is their modularity; due to their small size CBMs can be designed in tandem to increase specificity or allow for multiple binding targets. Additionally, there is a variety of well characterized CBMs that can be used as scaffolds—not surprisingly, CBMs have been used to engineer a variety of GBPs with altered binding characteristics[13][15][16][17].
4. Pseudoenzymes
In nature, a number of GBPs have evolved from enzymes through the loss of catalytic activity while retaining binding function. These can be defined as pseudoenzymes, which are catalytically inactive proteins related to ancestral enzymes[18]. Pseudoglycosidases are a type of pseudoenzyme that evolved from glycosidases (glycoside hydrolases). These proteins, which bind glycans but cannot hydrolyze glycosidic linkages, can also be characterized as lectins since glycan-binding is their primary function. A few notable examples of pseudoglycosidases that act as GBPs have been observed in nature. In animals, chitinase-like proteins such as the human YKL-39 are pseudoglycosidases (GH18 homologues) with enigmatic biological functions that have been shown to bind to chitooligosaccharides as part of their apparent role in modulating the innate immune response[19][20]. Another example in animals is found in α- and β-klotho proteins, which each make up part of a receptor complex responsive to fibroblast growth factors (FGFs), wherein catalytically inactive GH1-like tandem repeats of the klotho proteins bind to “sugar-mimicking motifs” of FGF19 and FGF21[21]. In protozoans, the CyRPA protein of Plasmodium falciparum—part of the invasion complex that allows the malaria-causing parasite to bind and enter red blood cells—appears to be a catalytically inactive pseudoglycosidase related to GH33 sialidases[22][23][24]. Pseudoenzymes evolved from other types of enzymes can also bind to glycans. For example, PgaB in E. coli is a deacetylase that is involved in the formation of the partially deacetylated poly-1,6-N-acetylglucosamine component of the bacterium’s biofilm coat, and the protein consists of two tandem domains related to carbohydrate esterase family 4 (CE4), with the C-terminal domain being a catalytically inactive pseudoesterase involved in binding poly-1,6-N-acetylglucosamine [25].
Although pseudoenzymes can, in theory, be used as glycan binding scaffolds, there are no published works on engineering pseudoenzyme scaffolds into novel GBPs as of 2020. This may be due to a lack of known pseudoenzymes scaffolds but may also be due to the prevalence of mutagenesis techniques that allow for inactivation of enzymes. The use of enzymes as GBP scaffolds is discussed in greater detail in the following section.
5. Carbohydrate-Active Enzymes (CAZymes)
Carbohydrate-active enzymes (CAZymes) catalyze reactions that break down, assemble, or modify saccharides, and they are categorized based on their activities and further subdivided into families based on sequences. Categories include glycosyltransferase, glycoside hydrolase, polysaccharide lyase, carbohydrate esterase, and auxiliary activity families. The examples of pseudoenzymes from nature demonstrate that inactivation of CAZymes can result in proteins that bind to glycans but do not catalytically turn them over. Naturally, this suggests that inactivating CAZymes through artificial mutations may be an effective method to engineer novel GBPs. Generating a GBP from a CAZyme requires the inactivation of catalytic residues, which sometimes only requires the mutation of a single amino acid. This may make CAZymes an attractive scaffold for GBP engineering. An example of a nanomolar affinity GBP engineered by inactivation of a CAZyme can be seen in the site-specific mutation of a glycoside hydrolase from E. coli K1 bacteriophages. The GH58 endosialidase, Endo-NF, was mutated to generate a catalytically inactive GBP that still binds to polysialic acid with a dissociation constant (KD) of 191 nM [26][27]. This engineered GBP has been applied as a very sensitive tool for detecting polysialic acid[28][29]. In another example, mutation of a CE2 carbohydrate esterase from Clostridium thermocellum has also been shown to produce a catalytically inactive GBP with micromolar affinity[30]. A single amino acid replacement of the CtCE2 enzyme not only abolished esterase activity, but increased the affinity to cellooligosaccharides nearly 8-fold, with the mutant binding to cellohexaose with a KD of 4.1 μM.
The strategy of engineering GBPs by inactivating CAZymes has been developed and commercialized most notably by the biotech company Lectenz Bio, who have produced a variety of catalytically inactivated CAZymes, which they have dubbed “Lectenz®” (lectins engineered from enzymes) [31]. The company has produced several Lectenz® through site-directed mutagenesis and computationally guided directed evolution. One advantage of using CAZyme scaffolds is that carbohydrate-processing enzymes tend to be more specific for their ligands than lectins, although this will vary between proteins.
6. Antibody-Based Scaffolds
Antibody-based scaffolds consist of immunoglobulin or immunoglobulin-like protein folds. A variety of antibody-based scaffolds are found in animals, but the most commonly used for developing antigen binding proteins are immunoglobulin G (IgG), and more recently, camelid antibodies[32]. The production of naturally occurring antibodies is time consuming and costly as it requires the immunization of an animal; however, antibody-based scaffolds have been engineered that circumvent the use of animals. These include, but are not limited to, antigen binding fragments (Fab)[33], single chain variable fragments (ScFvs)[34], diabodies[35], monobodies, and nanobodies [36]. There has been a concerted effort to produce antibodies against tumor-associated carbohydrate antigens (TACAs)—in total, antibodies have been designed for about 250 distinct glycan targets [37]. Antibody scaffolds offer certain advantages over lectins, including a larger binding interface for longer glycan epitopes, and generally more selective binding due to the complementary determining regions. However, glycans are poorly immunogenic and producing an anti-glycan antibody can be costly, labour intensive, and time consuming. Additionally, anti-glycan antibodies generally have lower affinities (KD in the micromolar range) than protein-targeting antibodies (KD in the nanomolar range). Within the last decade, phage display has provided methods for overcoming some of these limitations, resulting in antibodies with higher affinity for their glycan targets[38]. However, this approach still requires an initial scaffold obtained from immunization to be used as the base scaffold for improving affinity and selectivity.
7. Summary on Available GBP Scaffolds
Here we discussed the available protein scaffolds and some of their respective challenges and considerations when applied to GBP engineering. The scaffold that is chosen for GBP engineering will influence which mutagenesis techniques and selection methods are most appropriate. This brief overview provides a resource for glycobiologists who aim to design novel GBPs for specific glycan targets. Table 1 is by no means a complete scaffold list; it serves as a list of example scaffolds that are available and characteristics that need to be taken into consideration. Finding scaffolds ideal for a glycan of choice can be challenging and we recommend using UniLectin3D or equivalent GBP databases as starting point for finding potential scaffolds [9].
Table 1. Carbohydrate binding proteins (CBPs) with available structural and ligand binding information.
| Scaffold Category |
Scaffold Sub-Category |
Description | Origin | Example Protein (PE) | PE Length | PE Ligand | PE Oligomeric State | PE Multivalency |
|---|---|---|---|---|---|---|---|---|
|
Lectins |
P-type |
Lectin that binds to mannose 6-phosphate |
Animal |
Bovine CD-MPR binding domain [56] |
154 aa |
Mannose 6-Phosphate |
Dimer |
Monovalent |
|
I-type |
Protein that is homologous to the immunoglobulin superfamily (IgSF) |
Vertebrata |
hCD22 domains 1-3 [57] |
324 aa |
Sialoglycans |
Monomer |
Monovalent |
|
|
L-type |
Proteins that are structurally similar to lectins found in the seeds of leguminous plants |
All domains of life and viruses |
237 aa |
Trimannoside containing-oligosaccharides [59] |
Oligomer |
Divalent |
||
|
R-type |
Proteins that are structurally similar to the carbohydrate recognition domain (CRD) in ricin |
All domains of life and viruses |
Ricin [60] |
267 aa |
β1,4 galactose, N-acetylgalactosamine |
Dimer |
Divalent |
|
|
C-type |
Ca2+ dependant proteins that share a primary and secondary homology in their CRDs |
Animal |
C-type domain of murine DCIR2 [61] |
129 aa |
N-glycans |
Monomer |
Monovalent |
|
|
Galectin |
Globular proteins that share primary structural homology in their CRDs |
Animal |
hGalectin-3 [62] |
146 aa |
N-acetyllactosamine |
Monomer |
Monovalent |
|
|
Carbohydrate Binding Modules (CBMs) |
Type A |
Protein domain that binds to crystalline surfaces of cellulose and chitin |
All domains of life and viruses |
CBM from Cel7A [63] |
36 aa |
Cellulose |
Monomer |
Monovalent |
|
Type B |
Protein domain that binds endo-glycan chains |
All domains of life and viruses |
CBM4-2 from xylanase [33] |
150 aa |
Xylans, β-glucans |
Monomer |
Monovalent |
|
|
Type C |
Protein domain that binds exo-type glycan chains |
All domains of life and viruses |
Cp-CBM 32 of hexosaminidase [64] |
150 aa |
N-acetyllactosamine |
Monomer |
Monovalent |
|
|
Pseudoenzymes |
Pseudoglycosidase |
Carbohydrate binding proteins that evolved from glycosidases but are no longer catalytically active |
Possibly all domains of life* |
hYKL-39 [36] |
365 aa |
Chitooligo-saccharides |
Monomer |
Monovalent |
|
Pseudoesterase |
Carbohydrate binding proteins that evolved from carbohydrate esterases but are no longer catalytically active |
Possibly all domains of life * |
C-terminal domain of PgaB [42] |
367 aa |
Poly-1,6-N-acetylgluco-samine |
Monomer |
Monovalent |
|
|
Carbohydrate- Active Enzymes (CAZymes) |
Glycoside hydrolase |
Enzymes that cleave glycosidic linkages |
All domains of life and viruses |
Endo-NF (GH58) [44] |
811 aa |
Polysialic acid |
Trimer |
Multivalent |
|
Carbohydrate esterase |
Enzymes that hydrolyze ester linkages of acyl groups attached to carbohydrates |
All domains of life and viruses |
CtCE2 [47] |
333 aa |
Cellooligo-saccharides |
Monomer |
Monovalent |
|
|
Other CAZymes (glycosyltransferase, polysaccharide lyase, auxiliary activities) |
Enzymes involved in the assembly, break-down, and modification of carbohydrates |
All domains of life and viruses |
− |
− |
− |
− |
− |
|
|
Antibodies |
N/A |
Naturally or synthetically produced proteins with an immunoglobulin, or derived from an immunoglobulin-like structure |
Vertebrata |
hu3S193 [65] |
LC: 219 aa HC: 222 aa |
LewisY |
Dimeric |
Divalent |
References
- Barondes, S.H. Bifunctional properties of lectins: Lectins redefined. Trends Biochem. Sci. 1988, 13, 480–482, doi:10.1016/0968-0004(88)90235-6.
- Kocourek, J.; Horejsi, V. Defining a lectin. Nat. Cell Biol. 1981, 290, 188, doi:10.1038/290188a0.
- Feng, Y.; Guo, Y.; Li, Y.; Tao, J.; Ding, L.; Wu, J.; Ju, H. Lectin-mediated in situ rolling circle amplification on exosomes for probing cancer-related glycan pattern. Anal. Chim. Acta 2018, 1039, 108–115, doi:10.1016/j.aca.2018.07.040.
- Hashim, O.H.; Jayapalan, J.J.; Lee, C.-S. Lectins: An effective tool for screening of potential cancer biomarkers. PeerJ 2017, 5, e3784, doi:10.7717/peerj.3784.
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Preste-gard, J.H.; et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015.
- Bertozzi, C.R. Chemical Glycobiology. Science 2001, 291, 2357–2364, doi:10.1126/science.1059820.
- Liener, I.E.; Sharon, N.; Goldstein, I.J. The Lectins: Properties, Functions, and Applications in Biology and Medicine; Academic Press: Cambridge, MA, USA, 1986.
- Hu, D.; Tateno, H.; Hirabayashi, J. Lectin Engineering, a Molecular Evolutionary Approach to Expanding the Lectin Utili-ties. Molecules 2015, 20, 7637–7656, doi:10.3390/molecules20057637.
- Bonnardel, F.; Mariethoz, J.; Salentin, S.; Robin, X.; Schroeder, M.; Perez, S.; Lisacek, F.; Imberty, A. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 2019, 47, D1236–D1244, doi:10.1093/nar/gky832.
- Armenta, S.; Moreno-Mendieta, S.; Sánchez-Cuapio, Z.; Sanchez, S.; Rodríguez-Sanoja, R. Advances in molecular engineer-ing of carbohydrate-binding modules. Proteins Struct. Funct. Bioinform. 2017, 85, 1602–1617, doi:10.1002/prot.25327.
- Guillén, D.; Sanchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Micro-biol. Biotechnol. 2009, 85, 1241–1249, doi:10.1007/s00253-009-2331-y.
- Simpson, H.D.; Barras, F. Functional Analysis of the Carbohydrate-Binding Domains of Erwinia chrysanthemi Cel5 (En-doglucanase Z) and an Escherichia coli Putative Chitinase. J. Bacteriol. 1999, 181, 4611–4616, doi:10.1128/jb.181.15.4611-4616.1999.
- Simpson, P.J.; Jamieson, S.J.; Hachem, M.A.; Karlsson, E.N.; Gilbert, H.J.; Holst, O.; Williamson, M.P. The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. Biochemistry 2002, 41, 5712–5719, doi:10.1021/bi012093i.
- Boraston, A.B.; Creagh, A.L.; Alam, M.; Kormos, J.M.; Tomme, P.; Haynes, C.A.; Warren, R.A.J.; Kilburn, U.G. Binding speci-ficity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochem-istry 2001, 40, 6240–6247, doi:10.1021/bi0101695.
- Furtado, G.P.; Lourenzoni, M.R.; Fuzo, C.A.; Fonseca-Maldonado, R.; Guazzaroni, M.-E.; Ribeiro, L.F.; Ward, R.J. Engineer-ing the affinity of a family 11 carbohydrate binding module to improve binding of branched over unbranched polysaccha-rides. Int. J. Biol. Macromol. 2018, 120, 2509–2516, doi:10.1016/j.ijbiomac.2018.09.022.
- Gunnarsson, L.C.; Karlsson, E.N.; Albrekt, A.; Andersson, M.; Holst, O.; Ohlin, M. A carbohydrate binding module as a di-versity-carrying scaffold. Protein Eng. Des. Sel. 2004, 17, 213–221, doi:10.1093/protein/gzh026.
- Sakata, T.; Takakura, J.; Miyakubo, H.; Osada, Y.; Wada, R.; Takahashi, H.; Yatsunami, R.; Fukui, T.; Nakamura, S. Improve-ment of binding activity of xylan-binding domain by amino acid substitution. Nucleic Acids Symp. Ser. 2006, 50, 253–254, doi:10.1093/nass/nrl126.
- Eyers, P.A.; Murphy, J.M. The evolving world of pseudoenzymes: Proteins, prejudice and zombies. BMC Biol. 2016, 14, 1–6, doi:10.1186/s12915-016-0322-x.
- Schimpl, M.; Rush, C.L.; Betou, M.; Eggleston, I.M.; Recklies, A.D.; Van Aalten, D.M. Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide-binding properties. Biochem. J. 2012, 446, 149–157, doi:10.1042/bj20120377.
- Lee, C.G.; Da Silva, C.A.; Cruz, C.S.D.; Ahangari, F.; Ma, B.; Kang, M.-J.; He, C.-H.; Takyar, S.; Elias, J.A. Role of Chitin and Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annu. Rev. Physiol. 2011, 73, 479–501, doi:10.1146/annurev-physiol-012110-142250.
- Lee, S.; Choi, J.; Mohanty, J.; Sousa, L.P.; Tome, F.; Pardon, E.; Steyaert, J.; Lemmon, M.A.; Lax, I.; Schlessinger, J. Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nat. Cell Biol. 2018, 553, 501–505, doi:10.1038/nature25010.
- Chen, L.; Xu, Y.; Wong, W.; Thompson, J.K.; Healer, J.; Goddard-Borger, E.D.; Lawrence, M.C.; Cowman, A.F. Structural ba-sis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA. eLife 2017, 6, 213, doi:10.7554/elife.21347.
- Favuzza, P.; Guffart, E.; Tamborrini, M.; Scherer, B.; Dreyer, A.M.; Rufer, A.C.; Erny, J.; Hoernschemeyer, J.; Thoma, R.; Schmid, G.; et al. Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhib-itory antibody. eLife 2017, 6, doi:10.7554/eLife.20383.
- Wong, W.; Huang, R.; Menant, S.; Hong, C.; Sandow, J.J.; Birkinshaw, R.W.; Healer, J.; Hodder, A.N.; Kanjee, U.; Tonkin, C.J.; et al. Structure of Plasmodium falciparum Rh5–CyRPA–Ripr invasion complex. Nat. Cell Biol. 2018, 565, 118–121, doi:10.1038/s41586-018-0779-6.
- Little, D.J.; Li, G.; Ing, C.; DiFrancesco, B.R.; Bamford, N.C.; Robinson, H.; Nitz, M.; Pomès, R.; Howell, P.L. Modification and periplasmic translocation of the biofilm exopolysaccharide poly-1,6-N-acetyl-D-glucosamine. Proc. Natl. Acad. Sci. USA 2014, 111, 11013–11018, doi:10.1073/pnas.1406388111.
- Stummeyer, K.; Dickmanns, A.; Mühlenhoff, M.; Gerardy-Schahn, R.; Ficner, R. Crystal structure of the polysialic acid–degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 2004, 12, 90–96, doi:10.1038/nsmb874.
- Jakobsson, E.; Jokilammi, A.; Aalto, J.; Ollikka, P.; Lehtonen, J.V.; Hirvonen, H.; Finne, J. Identification of amino acid resi-dues at the active site of endosialidase that dissociate the polysialic acid binding and cleaving activities in Escherichia coli K1 bacteriophages. Biochem. J. 2007, 405, 465–472, doi:10.1042/bj20070177.
- Yu, C.-C.; Huang, L.-D.; Kwan, D.; Wakarchuk, W.; Withers, S.G.; Lin, C.-C. A glyco-gold nanoparticle based assay for α-2,8-polysialyltransferase from Neisseria meningitidis. Chem. Commun. 2013, 49, 10166–10168, doi:10.1039/c3cc45147j.
- Yu, C.-C.; Hill, T.; Kwan, D.; Chen, H.-M.; Lin, C.-C.; Wakarchuk, W.; Withers, S.G. A plate-based high-throughput activity assay for polysialyltransferase from Neisseria meningitidis. Anal. Biochem. 2014, 444, 67–74, doi:10.1016/j.ab.2013.09.030.
- Montanier, C.; Money, V.A.; Pires, V.M.R.; Flint, J.E.; Pinheiro, B.A.; Goyal, A.; Prates, J.A.M.; Izumi, A.; Stålbrand, H.; Mor-land, C.; et al. The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions. PLoS Biol. 2009, 7, e1000071, doi:10.1371/journal.pbio.1000071.
- Woods, R.J.; Yang, L. Glycan-Specific Analytical Tools. U.S. Patent 9,926,612, 27 March 2018.
- Arbabi-Ghahroudi, M. Camelid Single-Domain Antibodies: Historical Perspective and Future Outlook. Front. Immunol. 2017, 8, 1589, doi:10.3389/fimmu.2017.01589.
- Holliger, P.; Prospero, T.; Winter, G. Diabodies: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. USA 1993, 90, 6444–6448, doi:10.1073/pnas.90.14.6444.
- Nelson, A.L. Antibody fragments. mAbs 2010, 2, 77–83, doi:10.4161/mabs.2.1.10786.
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. ScFv Antibody: Principles and Clinical Applica-tion. J. Immunol. Res. 2012, doi:10.1155/2012/980250. Available online: https://www.hindawi.com/journals/jir/2012/980250/ (accessed on 2 January 2021).
- Sha, F.; Salzman, G.; Gupta, A.; Koide, S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017, 26, 910–924, doi:10.1002/pro.3148.
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Com-munity Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783, doi:10.1021/acschembio.6b00244.
- Stewart, A.; Liu, Y.; Lai, J.R. A strategy for phage display selection of functional domain-exchanged immunoglobulin scaf-folds with high affinity for glycan targets. J. Immunol. Methods 2012, 376, 150–155, doi:10.1016/j.jim.2011.12.008.




