| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Márió Gajdács | + 3228 word(s) | 3228 | 2021-01-13 04:14:38 | | | |
| 2 | Márió Gajdács | Meta information modification | 3228 | 2021-01-18 19:21:35 | | | | |
| 3 | Márió Gajdács | Meta information modification | 3228 | 2021-01-18 19:23:09 | | | | |
| 4 | Karina Chen | Meta information modification | 3228 | 2021-01-21 11:19:21 | | |
Video Upload Options
Pseudomonas aeruginosa is the most frequent cause of infection among non-fermenting Gram-negative bacteria, predominantly affecting immunocompromised patients, but its pathogenic role should not be disregarded in immunocompetent patients. seudomonas aeruginosa is the most frequent cause of infection among non-fermenting Gram-negative bacteria, predominantly affecting immunocompromised patients, but its pathogenic role should not be disregarded in immunocompetent patients.
1. Taxonomy and phenotypic characteristics of P. aeruginosa
Based on phenotypic characteristics, Gilardi has classified NFGNB into seven main groups, while Palleroni has differentiated five distinct homologous rRNA groups (namely, I.: Pseudomonas, II.: Burkholderia, III.: Comamonas, IV.: Brevimundas, and V.: Stenotrophomonas) based on rRNA-DNA homology [1][2]. P. aeruginosa was first isolated from green pus by Gessard in 1882, while the genus Pseudomonas was first described by Migula in 1894, with P. aeruginosa being the species type of the genus [9]. Members of the Pseudomonaceae family are ubiquitous in nature (soil, plants, and aquatic environments, while birds and smaller mammals have also been described as reservoirs) [3][4][5]. P. aeruginosa is the most common cause of infections (both within the genus and among NFGNB) in humans and warm-blooded animals (e.g., urinary tract infections, mastitis, and endometritis in livestock and companion animals) [6]. Other members of the genus are relevant as fish pathogens, causing hemorrhagic septicemia and ulcerative syndrome [7][8]. P. fluorescens and P. putida have been described as a cause for deterioration of refrigerated food, and as contaminants in blood transfusion and infusion preparations. P. stutzeri, P. mendocina, P. fulva, and P. monteilii are rarely pathogenic in humans (described in patients with end-stage disease and in septicemia), while P. baetica, P. syringae, P. plecoglossicida, and P. viridiflava are important plant pathogens [9][10][11].
P. aeruginosa is a non-fastidious microorganism that does not require special cultivation conditions. It grows well on most non-selective (Mueller-Hinton, Nutrient agar, Luria-Bertani, blood agar, etc.) media, although there are some media which are used specifically for the purpose of selective propagation of Pseudomonas (e.g., cetrimide agar, King-A, and King-B media). While the microorganism grows best at 37 °C, pseudomonads can also survive in a wide temperature range (4–40 °C) [12][13]. Among the phenotypic characteristics of P. aeruginosa, the characteristic odor (described as flower-like, “grape juice”, or “fresh tortilla”), β-haemolysis (on blood agar), and color of the colonies (in appropriate culture media) allows for their quick organoleptic identification [14]. P. aeruginosa and other members of the genus are known to produce various pigments, including pyoverdine/fluoresceine (a fluorescent green-yellow water-soluble pigment, produced by 70–80% of isolates, which also acts as a siderophore in low-iron conditions), pyocyanin (a green-blue lipid-soluble phenazone-derivative pigment, with roles in iron metabolism, in maintaining the redox-equilibrium surrounding the bacteria, and in cell–cell communication), pyorubin (a red-brown water-soluble pigment, produced by 2–3% of isolates, with roles in maintaining the redox-equilibrium), and pyomelanin/alkaptomelanin (a brown-black, water-soluble, and acidic pigment) [15][16][17][18] (Figure 1). It has been shown that high phosphate concentration in the culture media induces pigment production in Pseudomonas spp. [19][20].
Figure 1. P. aeruginosa antimicrobial susceptibility testing using disk diffusion on Mueller-Hinton agar plates. The isolate on the upper portion of the figure produces pyorubin and pyomelanin, while the isolate on the lower portion of the figure produces pyocyanin.
2. Virulence Determinants of P. aeruginosa and Modulation of Virulence Factor Expression
2.1. Genome of P. aeruginosa
The pathogenicity of P. aeruginosa is supported by numerous virulence determinants, some of which are integral parts of their cell structure. On the other hand, many additional virulence factors are synthesized and excreted, depending on the environment surrounding the pathogen [21][22]. One of the most important characteristics of P. aeruginosa is its adaptability to diverse natural environments and to harsh (in vivo) conditions, which concurs with high metabolic diversity among species of this genus [23]. The publication of the first sequenced genome of the opportunistic pathogenic strain P. aeruginosa PAO1 (isolated from a wound) by Stover et al. in 2000 had paramount importance in shedding light on the physiology and virulence capabilities of this pathogen [24][25]. Since then, the complete genome of many other species of the genus (P. putida KT2440, P. fluorescens Pf-5, P. fluorescens PfO-1, P. entomophila L48, and others) have been published [24][25]. In comparison with a common Gram-negative isolate, namely the uropathogenic Escherichia coli (UPEC) (with a genome of ≥5 Mb) [26][27], P. aeruginosa has a large genome with 5.5–7 Mb, characterized by pronounced genomic plasticity [28][29]. This genetic repertoire includes a conserved core genome of ~4 Mb, while the remaining genetic material comprises of various sets of rare genes and gene islands [30]. The versatility of this pathogen is largely determined by the latter group of genes.
The P. aeruginosa genome resembles a classical “secretor” genome, which includes a large proportion of regulatory genes (i.e., efflux pumps and other transport proteins, motility, chemotaxis), genes controlling metabolic pathways (which allows for adapting to distinct metabolic states), and genes encoding a plethora of virulence factors and antibiotic resistance determinants [38,39]. For example, cystic fibrosis—the defect of the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) genes—leads to the accumulation of succinate in the lungs, which favors the colonization and survival of P. aeruginosa, as this microorganism can utilize it as a nutrient source [40]. Secreted virulence factors and proteases are some of the hallmarks in P. aeruginosa pathogenicity, which take up ~3% of the open reading frames of the P. aeruginosa PAO1 genome [24][25]. The diversity of the P. aeruginosa genome is further enhanced by the introduction of mobile genetic elements via horizontal gene transfer (HGT; such as conjugative transposons, insertion sequences, and genomic islands) [31]. P. aeruginosa also has an innate way to increase genetic diversity in hypermutable strains: the DNA-mismatch repair system in these microorganisms consist of a protein trimer (namely the MutS-MutL-UvrD trimer), with the role of maintaining genomic integrity in these species [32][33]. It has been suggested that species with mutations in this repair system result in “hypermutator” strains, where the spontaneous mutation rate is increased 1000×. These isolates are principally seen in the lung of CF patients and they are characterized by phenotypic changes (i.e., the so-called “mucoid” phenotype) and high-level antibiotic resistance [34][35].
2.2. Virulence Factors of P. aeruginosa
Similar to other Gram-negative bacteria, lipopolysaccharide (LPS, or endotoxin), Type IV pili and flagella, adhesins, and lectins are all integral parts of the external cell wall structure of P. aeruginosa [36][37]. Based on the O-specific polysaccharide side chain of the LPS, 27 antigen groups may be differentiated, while there is also an opportunity to classify these bacteria based on their flagellar H-antigens [38]. The feature of motility for P. aeruginosa is recognized as an advantage, as it is able to move from one niche to another with no difficulty [36][37][39]. Three types of motility, including swarming, swimming, and twitching motility, enable P. aeruginosa to be present in a wide range of different habitats with a diversity of environmental factors [40]. Lectins are proteins on the outer membrane of P. aeruginosa, which recognize glycosylated carbohydrates on host tissues, aiding the adherence of bacterial cells. For example, LecA (which binds to galactose) and LecB (which binds to fucose) mediate the adherence of this pathogen to epithelial cells in the lung [41][42]. These cell-mediated virulence determinants have important roles in the initial phase of colonization, persistence, and in the establishment of infections in vivo [43]. Nevertheless, the overwhelming majority of virulence determinants associated with P. aeruginosa are secreted factors. These may be synthesized and secreted to the vicinity of these bacteria (damaging surrounding tissues and immune cells). In addition, they may be introduced directly into host cells via a type III secretion system (T3SS) [44][45][46]. Secreted virulence factors are relevant in the later stages of the infection and invasion, during which bacterial cells proliferate and subsequent damage occurs in tissue cells at the anatomical site of infection, and the host immune response is dampened [47].
These secreted virulence factors in P. aeruginosa include: (i) pigments (described previously), siderophores (e.g., achromobactin), and inorganic compounds (e.g., hydrogen cyanide), which have roles in iron scavenging, protection against damage caused by reactive oxygen species (ROS; originating from immune cells), and competition against other bacterial genera. (ii) Exotoxins, including effector cytotoxins such as exotoxin A (ETA), exotoxin S (ExoS; inhibits the function of innate immune cells and neutrophil granulocytes), exotoxin U (ExoU; phospholipase activity, which rapidly leads to cell lysis and has roles in inducing septic shock), and other exotoxins with similar functions (ExoT: inhibits cell division in mammalian cells and affects wound healing processes, ExoY: induces pro-apoptotic processes, and exolysin A (ExlA), which is secreted by a two-partner secretion system (TPS)). (iii) Proteases and other enzymes: lipases, alkaline protease, elastase A (LasA), and B (LasB), heat-stable hemolysin/phospholipase H (PLH), phospholipase C (PLC), and DNase. (iv) Secretion systems: P. aeruginosa is known to have 5 types of secretion systems, among which, Types I (T1SS), II (T2SS), and III are involved in the virulence of this pathogen. T1SS and T2SS are relevant in the secretion of various proteases and lipases, ETA, LasA, LasB, and PLH. On the other hand, there are two distinct T3SSs in P. aeruginosa: the role of fT3SS is to expel flagellar proteins (to aid in motility, and they may also play a role in biofilm formation), while the iT3SS is a needle-like protein (“injectasome”), which introduces the previously mentioned effector toxins (such as ExoU and ExoS) into the cytoplasm of mammalian cells [44][45][46][48][49][50][51][52][53][54]. (v) Biofilm (see Section 2.4). In contrast to cell-mediated virulence factors (which are considered to be constitutive), the production of secreted virulence factors is largely dependent on the environmental factors and the niche surrounding the pathogen.
2.3. Typing methods for the differentiation of P. aeruginosa clones, global dissemination
Many methods (with various costs, labor-intensity, and discriminatory power) have been proposed for the assessment of genetic similarity in P. aeruginosa, which are just as important for local infection control interventions and outbreak control as they are relevant in the assessment of successful national or global clones by public health microbiology [55][56]. These methods include serotyping, phage typing (based on the differential sensitivity to these isolates to standardized bacteriophages), pyocin typing, pulse-field gel electrophoresis (PFGE), field-inversion gel electrophoresis (FIGE), random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR), oligonucleotide microarrays, multi-locus sequence typing (MLST), and whole-genome sequencing (WGS) [57][58][59]. While the latter three methods are relevant in the identification of internationally successful P. aeruginosa clones, the other listed typing methods are used in the assessment of local outbreaks. Currently, three major international multidrug-resistant (MDR) clones have been identified, which have shown the most successful spread around the globe, namely the ST111, ST175, and ST235 clones [69,70]. ST111 (characterized by serotype O12) and ST235 (characterized by serotype O11) have been described on almost every continent of the world, while ST175 (characterized by serotype O4) has only been detected in European countries [60][61].
ST235 clones are known as highly virulent—owing to the presence of ExoU in these strains—and these isolates are MDR; thus, the therapy of these infections is also considerably more difficult. Generally, it may be said that the continuous expression of resistance-determinants hinders the virulence of the microorganism; however, the fitness burden associated with maintaining the MDR-phenotype was observed to be lower in case of the ST235 clones [62]. Based on WGS analysis, P. aeruginosa isolates of clinical and environmental origin may be grouped into three distinct resistotypes, namely PAO1, PA14, and PA7. PAO1 and PA14 are characterized by possessing the T3SS secretion system and the corresponding effector toxins (ExoS but not ExoU in the case of PAO1, and ExoU but not ExoS in the case of PA14). On the other hand, PA7 does not have the T3SS; instead, they utilize the TPS, by which they secrete the ExlA exolysin to damage surrounding tissue cells [63]. Some reports suggest that there may be an association between virulence and antibiotic resistance in P. aeruginosa isolates, as the carriage of the exoU genes was shown to correlate with resistance to aminoglycosides and fluoroquinolones. A possible explanation was that the genomic island carrying exoU may also contain resistance-determinant genes [64][65].
2.4. Biofilm Formation
Without a doubt, one of the most important virulence determinants in the pathogenesis of P. aeruginosa infections is the production of a biofilm. The biofilm allows for the adherence of these pathogens on various surfaces, provides protection from harsh environmental conditions (e.g., sheer forces, drying), and from the immune system of the host (e.g., natural killer cells, phagocytes, complement, ROS-mediated damage) [66][67][68]. Biofilms have heterogenous compositions, consisting of aggregates of sessile bacterial communities (based on their composition, this may be monospecies or multispecies biofilm), exopolysaccharides (EPS; e.g., alginate, cellulose, dextran, rhamnolipids), environmental DNA (eDNA), carbohydrates, proteins, surfactants, lipids, various ions, and water [69][70]. The biofilm mode of growth was first described in the 1930s, while the true relevance of biofilm-embedded bacteria in infectious processes has been understood only in recent decades [71][72]. Bacterial cells usually attach to hydrophobic and/or coarse surfaces with the aid of their cell-mediated virulence determinants (e.g., pili, fimbriae, surface antigens), which is followed by the production of the protective EPS and other components [73]. Biofilms allow P. aeruginosa to persist in the external environment (in water pipes and tanks, sinks, on hospital tiles, on medical equipment, such as mechanical ventilators and respiratory tubing, humidifiers, dialysis equipment and catheters, endoscopes and implanted medical devices, in medical preparations, such as irrigation solutions, dialysis fluid, contact lens fluid, antiseptic solution, cremes) and in vivo [74][75].
Biofilm formation is a critical attribute of P. aeruginosa in being a successful nosocomial pathogen and it is also an important hallmark of chronic bacterial persistence. This may be observed in dental caries on the tooth surfaces [76][77], in skin and soft tissue infections [78], in infections of the middle ear [87], catheter-associated infections [79], pneumonia, and in the lungs of CF patients [80]. In the latter case, P. aeruginosa is able to survive and avoid clearance (withstanding the immune response and the subsequent administration of antimicrobials) in the respiratory and conductive zone of the lungs [81][82]. For example, alginate and other polysaccharides produced by the mucoid variants are effective in scavenging ROS, protecting bacterial cells [83]. Other than the protection against immune cells, the biofilm provides a safe haven for microorganisms against antibiotics in vivo, contributing to the MDR phenotype. It has been noted by several publications that the minimum inhibitory concentrations (MICs) of bacteria inside the biofilm may be 10–10,000 times higher, compared to planktonic cells [81][82][84]. On one hand, the secreted extracellular matrix significantly hinders the diffusion of the antibiotic molecules to effectively reach the bacterial targets (pharmacokinetic barrier); in addition to this, bacteria residing in the deeper layers of the biofilm will adapt to a differentiated metabolic state [81][82][84]. It must be noted that the inhibition of bacterial growth is mechanistically distinct from bacterial killing, and antimicrobials (even in effective doses) may not kill cells inside a biofilm. Due to the high bacterial density, low oxygen tension, and lack of nutrients, bacteria become dormant and utilize alternative metabolic pathways [83].
In addition to lacking cell motility, these “persister” cells (also termed small-colony variants (SCVs)) correspond to a transient phenotypic variant of bacteria, which are not genetically resistant to antibiotics, but under the abovementioned conditions, they can withstand very high concentrations of these drugs (pharmacodynamic barrier) [85][86][87][88]. In essence, persisters (corresponding to 1–2% of the bacterial population) opt not to proliferate during exposure to antibiotics, but they resume replication if the stressors are removed from the environment [85][86][87][88]. Persisters may also be important in the recurrence and chronicity of P. aeruginosa infections. Although there is scarce knowledge on the mechanisms leading to dormancy/persister formation, it has been suggested that the secretion systems may have a role [89]. The therapy of biofilm infections is an important challenge, as there is currently no targeted therapy available to completely eradicate biofilms in vivo. Nevertheless, several compounds (e.g., polyvalent anions, DNases like dornase-α and alginate lyase) may be useful in the reduction of mucus density [90]. On the other hand (although the evidence on this topic is still controversial), some experiments have shown that sub-MIC concentrations of some antibiotics (mainly β-lactams, including ceftazidime, cefepime, imipenem, and meropenem) may have the opposite effect, inducing biofilm production [91][92][93]. P. aeruginosa also displays the ability to tolerate biocides (e.g., antiseptics and disinfectants) like chlorhexidine or triclosan, mediated by the fabV gene, coding for a triclosan-resistant enoyl-acyl-carrier protein. Lack of susceptibility to biocides further hinders successful elimination of P. aeruginosa from hospital environments [94][95].
2.5. Quorum Sensing (QS)-Mediated Control of Virulence Factor Expression in P. aeruginosa
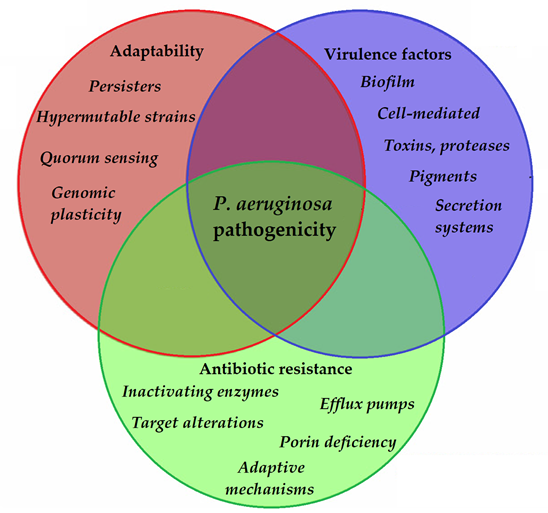
To allow for the continuous adaptation of P. aeruginosa to different environmental niches and to the different stages of infection, the secretion of the abovementioned virulence factors needs to be tightly regulated. One of the most important regulators in P. aeruginosa is by its quorum sensing (QS) systems [96]. QS corresponds to the “social behavior” of bacteria, during which small signal molecules (termed autoinducers) are used to influence gene expression in bacterial cells, in a cell-to-cell and density-dependent manner [97][98]. If the density of the bacterial population (hence, the concentration of these signal molecules) reaches a certain threshold, changes occur in bacterial physiology to aid collective behaviors or to help microorganisms to outcompete other microorganisms in the ecological niche (e.g., by secreting virulence factors or antibacterial compounds) [99][100]. Four interconnected systems, namely the iqs, las, pqs, and rhl pathways, compose the QS-regulatory network of Pseudomonas species. In this network, various autoinducers (such as acyl-homoserine lactones (acyl-HSLs), like butanoyl homoserine lactone (C4 HSL) and 3-oxodecanoyl homoserine lactone (C12 HSL)), the B. cepacia complex fatty acid molecule named diffusible signal factor (BDSF), oligopeptide-type autoinducers (like autoinducer-2 (AI-2)), the Pseudomonas quinolone signal molecule (PQS), and integrated QS signal molecule (IQS)) are utilized. The detailed description of these signal molecules is outside of the scope of this review (for details, see References [101][97][98][99][100][102][103][104][105][106][107]). Additionally, these autoinducer molecules are capable of dampening the innate immune response and inducing cytokines and chemokines [108]. As the production of biofilm and the secretion of other virulence factors are all governed by the complex QS system of P. aeruginosa, they have significant influence on the virulence of these bacteria. QS mediates the expression of its pigments, alkaline protease, hemolysin, elastase, lectins, the effector exotoxins, exotoxin A, swimming and twitching motilities, the activity of the T1SS and T2SS (the activity of T3SS is influenced by QS to a lesser extent), production of biofilm, and hydrogen cyanide, among others [109][110]. QS is also an important mediator of the reciprocity between bacterial virulence, antibiotic resistance, and microbial fitness [111]. The complexity of P. aeruginosa pathogenicity is represented in Figure 2.
Figure 2. Main components of P. aeruginosa pathogenicity.
Nonetheless, it is well-known that the upkeep of many resistance determinants and virulence factors may bear high fitness costs, leading to more susceptible strains outcompeting MDR ones [110][111]. Conversely, therapeutic and sub-inhibitory doses of antibiotics (e.g., ceftazidime, cefepime, imipenem, fluoroquinolones, doxycycline) may eradicate various QS signal molecules or inhibit their binding to the relevant receptors; thus, suppressing their virulence [112][113]. In addition to QS, biofilm production is also mediated by various two-component regulatory systems (GacS/GacA, RetS/LadS) and cyclic diguanylate (cyclic-di-GMP; a cyclic dimeric guanosine monophosphate), with the signal molecule having critical roles in the secretion of EPS [114]. If these bacteria adhere to any in vivo or inanimate surfaces, the concentration of cyclic-di-GMP increases, leading to the expression of “static” determinants, such as adhesive pili (securing attachment to the surface) and the subsequent production of biofilm. At the same time, increased cyclic-di-GMP also results in the repression of the synthesis and function of flagella (“motility” determinants) [114][115]. This leads to the thickening of the initial biofilm, corresponding to protection against immune cells and antimicrobials.
References
- Gilardi, G.L. Pseudomonas species in clinical microbiology. Mt. Sinai J. Med. 1976, 43, 710–726.
- Palleroni, N.J. Pseudomonas classification. A new case history in the taxonomy of gram-negative bacteria. Antonie Van Leeuwenhoek 1993, 64, 231–351.
- Gumey, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Pub. Health 2020, 1,148–157.
- Blanc, D.S.; Francioli, P.; Zanetti, G. Molecular Epidemiology of Pseudomonas aeruginosa in the Intensive Care Units—A Review. Open Microbiol. J. 2007, 1, 8–11.
- Klockgether, J.; Tömmler, B. Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F100Research 2017, 6, e1261.
- Maenni, M.; Bour, M.; Chatre, P.; Madec, J.Y.; Plésiat, P.; Jeannot, K. Resistance of Animal Strains of Pseudomonas aeru-ginosa to Carbapenems. Front. Microbiol. 2017, 8, e1841.
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H., El-Kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR‑Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX‑M, and tetA antibiotic‑resistance genes. Sci. Rep. 2020, 10, e15961.
- Ruiz-Roldán, L.; Rojo-Bezares, B.; de Toro, M.; López, M.; Toledano, P.; Lozano, C.; Chichón, G.; Alvarez-Erviti, L.; Torres, C.; Sáenz, Y. Antimicrobial resistance and virulence of Pseudomonas spp. among healthy animals: Concern about exolysin ExlA detection. Sci. Rep. 2020, 10, e11667.
- I.; Ramos, J.L.; et al. Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains. Front. Microbiol. 2015, 6, e871.
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vincente, A. Pseudomonas syringae pv. syringae Associated with Mango Trees, a Particular Pathogen Within the “Hodgepodge” of the Pseudomonas syringae Complex. Front. Plant Sci. 2019, 10, e570.
- Shariff, A.M.; Beri, K. Exacerbation of bronchiectasis by Pseudomonas monteilii: A case report. BMC Infect. Dis. 2017, 17, e511.
- Brown, V.I.; Lowbury, E.J.L. Use of an improved cetrimide agar medium and other culture methods for Pseudomonas aeruginosa. J. Clin. Pathol. 1965, 18, 752–756.
- Gajdács, M.; Burián, K.; Terhes, G. Resistance Levels and Epidemiology of Non-Fermenting Gram-Negative Bacteria in Urinary Tract Infections of Inpatients and Outpatients (RENFUTI): A 10-Year Epidemiological Snapshot. Antibiotics 2019, 8, e143.
- Clark, S.T.; Caballero, J.D.; Cheang, M.; Coburn, B.; Wang, P.W.; Donaldson, S.L.; Zhang, Y.; Liu, M.; Keshavjee, S.; Yau, Y.C.W.; et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci. Rep. 2015, 5, 10932.
- Behzadi, P.; Behzadi, E. The microbial agents of urinary tract infections at central laboratory of Dr. Shariati Hospital, Tehran, Iran. Turk. Klin. Tip Bilim. 2008, 28, e445.
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.J.; Forbes, A.; Perkis, A.V.; Davey, A.K.; Chess-Williams, R.; Kiefel, M.J.; Arora, D.; et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 2016, 8, e236.
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, A.I. Pyoverdin is essential for virulence of Pseudomonas aeru-ginosa. Infect. Immun. 1996, 64, 518–523.
- Alyydice-Francis, K.; Brown, P.D. Diversity of Antimicrobial Resistance and Virulence Determinants in Pseudomonas aeruginosa Associated with Fresh Vegetables. Int. J. Microbiol. 2012, 2012, e426241.
- El-Fouly, M.Z.; Sharaf, A.M.; Shahin, A.A.M.; El-Bialy, H.A.; Omara, A.M.A. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J. Rad. Res. Appl. Sci. 2015, 8, 36–48.
- Behzadi, P.; Behzadi, E. A study on apoptosis inducing effects of UVB irradiation in Pseudomonas aeruginosa. Roum. Arch. Microbiol. Immunol. 2011, 70, 74–77.
- Azuama, O.C.; Ortiz, S.; Quirós-Guerrero, L.; Bouffartigues, E.; Tortuel, D.; Maillot, O.; Feuilloley, M.; Cornelis, P.; Lesouhaitier, O.; Grougnet, R.; et al. Tackling Pseudomonas aeruginosa Virulence by Mulinane-Like Diterpenoids from Azorella atacamensis. Biomolecules 2020, 10, e1626.
- Shaw, E.; Wuest, W.M. Virulence attenuating combination therapy: A potential multi-target synergy approach to treat Pseudomonas aeruginosa infections in cystic fibrosis patients. RSC Med. Chem. 2020, 11, e358.
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680.
- Vodovar, N.; Vallenet, D.; Cruveiller, S.; Rouy, Z.; Barbe, V.; Acosta, C.; Cattolico, L.; Jubin, C.; Lajus, A.; Segurens, B.; et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas ento-mophila. Nat. Biotechnol. 2006, 24, 673–679.
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Lagrou, M.; Garber, R.L.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic patho-gen. Nature 2006, 406, 959–964.
- Hozzari, A.; Behzadi, P.; Khiabani, P.K.; Sholeh, M.; Sabokroo, N. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J. Appl Gen. 2020, 61, 265–273.
- Issakhanian, L.; Behzadi. P. Antimicrobial agents and urinary tract infections. Curr. Pharm. Des. 2019, 25, 1409–1423.
- de Lorenzo, V. Pseudomonas aeruginosa: The making of a pathogen. Environ. Microbiol. 2015, 17, 1–3.
- Suenaga, H.; Fujihara, H.; Kimura, N.; Hirose, J.; Watanabe, T.; Futagami, T.; Goto, M.; Shimodaira, J.; Furukawa, K. In-sights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and ge-nome instability properties. Environ. Microbiol. Rep. 2017, 9, 589–598.
- Rumbaugh, K.P. Genomic complexity and plasticity ensurePseudomonas success. FEMS Microbiol. Lett. 2014, 356, 141–143.
- Bonomo, R.A.; Szabó, D. Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56.
- Hogardt, M.; Heesemann, J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top. Microbiol. Immunol. 2013, 358, 91–118.
- Rees, V.E.; Lucas, D.S.D.; López-Causapé, C.; Huang, Y.; Kotsimbos, T.; Bulitta, J.B.; Rees, M.C.; Barugahare, A.; Peleg, A.Y.; Nation, R.L.; et al. Characterization of Hypermutator Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis in Australia. Antmicrob. Agents Chemother. 2019, 63, e02538–e18.
- Ciofu, O.; Fussing, V.; Bagge, N.; Koch, C.; Hřiby, N. Characterization of paired mucoid/non-mucoid Pseudomonas aeru-ginosa isolates from Danish cystic fibrosis patients: Antibiotic resistance, beta-lactamase activity and RiboPrinting. J. Anti-microb. Chemother. 2001, 48, 391–396.
- Lee, B.; Schjerling, C.K.; Kirkby, N.; Hoffmann, N.; Borup, R.; Molin, S.; Hoiby, N.; Coifu, O. Mucoid Pseudomonas aeru-ginosa isolates maintain the biofilm formation capacity and the gene expression profiles during the chronic lung infection of CF patients. APMIS 2011, 119, 263–274.
- Jain, R.; Behrens, A.J.; Kaever, V.; Kazmierczak, B.I. Type IV Pilus Assembly in Pseudomonas aeruginosa over a Broad Range of Cyclic di-GMP Concentrations. J. Bacteriol. 2012, 194, 4285–4294.
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocincová, D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2011, 2, e118.
- Pan, X.; Cui, X.; Zhang, F.; He, Y.; Li, L.; Yang, H. Genetic Evidence for O-Specific Antigen as Receptor of Pseudomonas aeruginosa Phage K8 and Its Genomic Analysis. Front. Microbiol. 2016, 7, e252.
- Pan, X.; Cui, X.; Zhang, F.; He, Y.; Li, L.; Yang, H. Genetic Evidence for O-Specific Antigen as Receptor of Pseudomonas aeruginosa Phage K8 and Its Genomic Analysis. Front. Microbiol. 2016, 7, e252.
- Kazmierczak, B.I.; Schnierderberend, M.; Jain, R. Cross-regulation of Pseudomonas motility systems: The intimate relationship between flagella, pili and virulence. Curr. Opin. Microbiol. 2015, 28, 78–82.
- Chemani, C.; Imberty, A.; de Bentzmann, S.; Pierre, M.; Wimmerová, M.; Guery, B.P.; Faure, K. Role of LecA and LecB Lectins inPseudomonas aeruginosa-InducedLung Injury and Effect of Carbohydrate Ligands. Infect. Immun. 2009, 77, 2065–2075.
- Thuenauer, R.; Landi, A.; Trefzer, A.; Altmann, S.; Wehrum, S.; Elerhoff, T.; Diedrich, B.; Dengjel, J.; Nyström, A.; Im-berty, A.; et al. The Pseudomonas aeruginosa Lectin LecB Causes Integrin Internalization and Inhibits Epithelial Wound Healing. mBio 2020, 11, e03260–e19.
- Gilboa-Garber, N.; Avichezer, D. Effects of Pseudomonas aeruginosa PA-I and PA-II Lectins on Tumoral Cells. In Lectins and Glycobiology; Gabius, H.J., Gabius, S., Eds.; Springer: Berlin, Germany, 1993.
- Szabó, D.; Szentandrássy, J.; Juhász, Z.; Katona, K.; Nagy, K.; Rókusz, L. Imported PER-1 producing Pseudomonas aeru-ginosa, PER-1 producing Acinetobacter baumannii and VIM-2 producing Pseudomonas aeruginosa strains in Hungary. Ann. Clin. Microbiol. Antimicrob. 2008, 7, e12.
- Vanderwoude, J.; Fleming, D.; Azimi, S.; Trivedi, U.; Rumbaugh, K.R.; Diggle, S.P. The evolution of virulence in Pseu-domonas aeruginosa during chronic wound infection. Proc. R. Soc. B 2020, 287, e20202272.
- Veesenmeyer, J.L.; Hauser, A.R.; Lisboa, T.; Rello, J. Pseudomonas aeruginosa virulence and therapy: Evolving transla-tional strategies. Crit. Care Med. 2009, 37, 1777–1786.
- Amsalu, A.; Sapula, S.A.; Lopes, M.D.B.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Eux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Dierent Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms 2020, 8, 1647.
- Amsalu, A.; Sapula, S.A.; Lopes, M.D.B.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Eux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Dierent Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms 2020, 8, e1647.
- Crousilles, A.; Maunders, E.; Bartlett, S.; Fan, C.; Emem-Fong, U.; Abdelhamid, Y.; Baker, Y.; Floto, A.; Spring, D.R.; Welch, M. Which microbial factors really are important in Pseudomonas aeruginosa infections? Future Microbiol. 2015, 10, 1825–1836.
- Cabot, G.; Ocampo-Sosa, A.A.; Domínguez, M.A.; Gago, J.F.; Juan, C.; Tubau, F.; Rodriguez, C.; Moyá, C.; Mar-tínez-Martínez, L.; Oliver, A. Spanish Network for Research in Infectious Diseases (REIPI). Genetic markers of wide-spread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012, 56, 6349–6357.
- Manos, J.; Hu, H.; Rose, B.R.; Wainwright, C.E.; Zablotska, I.B.; Cheney, J.; Turnbull, L.; Whitchurch, C.B.; Grimwood, K.; Harmer, C.; et al. Virulence factor expression patterns in Pseudomonas aeruginosa strains from infants with cystic fi-brosis. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1583–1592.
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011, 13, 3128–3138.
- Chung, J.C.S.; Rzhepishevska, O.; Ramstedt, M.; Welch, M. Type III secretion system expression in oxygen-limited Pseu-domonas aeruginosa cultures is stimulated by isocitrate lyase activity. Open Biol. 2013, 3, e120131.
- Yahr, T.L.; Wolfgang, M.C. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Mi-crobiol. 2006, 62, 631–640.
- Waters, V.; Zlosnik, J.E.A.; Yau, Y.C.W.; Speert, D.P.; Aaron, S.D.; Guttman, D.S. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3341–3350.
- Magalhaes, B.; Valot, B.; Abdelbary, M.H.H.; Prod’hom, G.; Greub, G.; Senn, L.; Blanc, D.S. Combining Standard Molec-ular Typing and Whole Genome Sequencing to Investigate Pseudomonas aeruginosa Epidemiology in Intensive Care Units. Front. Public Health 2020, 8, e3.
- Hernandes, J.; Ferrus, M.A.; Hernandez, M.; Alonso, J.L. Comparison of six different methods for typing Pseudomonas aeruginosa strains isolated from bottled and well waters. Water Res. 1997, 31, 3169–3174.
- Ballarini, A.; Scarlet, G.; Kos, M.; Cramer, N.; Wiehlmann, L.; Jousson, O. Molecular typing and epidemiological investi-gation of clinical populations of Pseudomonas aeruginosa using an oligonucleotide-microarray. BMC Microbiol. 2012, 12, e152.
- MacCannell, D. Bacterial strain typing. Clin. Lab. Med. 2013, 33, 629–650.
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Res. Update 2015, 21, 41–59.
- Treepong, P.; Kos, V.N.; Goyeux, C.; Blanc, D.S.; Bertand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266.
- Wi, Y.M.; Choi, J.Y.; Lee, J.Y.; Kang, C.I.; Chung, D.R.; Peck, K.R.; Song, J.H.; Ko, K.S. Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int. J. Antimicrob. Agents 2017, 49, 767–769.
- Reboud, E.; Basso, P.; Maillard, A.P.; Huber, P.; Attrée, I. Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers. Toxins 2017, 9, 364.
- Khan, M.; Stapleton, F.; Summers, S.; Rice, S.A.; Willcox, M.D.P. Antibiotic Resistance Characteristics of Pseudomonas aeruginosa Isolated from Keratitis in Australia and India. Antibiotics 2020, 9, e600.
- Subedi, D.; Vijay, A.K.; Kohli, G.S.; Rice, S.A.; Willcox, M. Association between possession of ExoU and antibiotic re-sistance in Pseudomonas aeruginosa. PLoS ONE 2018, 13, e0204936.
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacte-rial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11.
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Grosu, M.; Stavropoulos, E.; Chifiriuc, M.C.; Lazăr, V. Microbial Biofilms in Urinary Tract Infections and Prostatitis: Etiology, Pathogenicity, and Combating strategies. Pathogens 2016, 5, 65.
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439.
- Behzadi, P.; Urbán, E.; Gajdács, M. Association between Biofilm-Production and Antibiotic Resistance in Uropathogenic Escherichia coli (UPEC): An In Vitro Study. Diseases 2020, 8, e17.
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Boil. Rev. 2014, 78, 510–543.
- Gajdács, M.; Urbán, E. Epidemiological Trends and Resistance Associated with Stenotrophomonas maltophilia Bactere-mia: A 10-Year Retrospective Cohort Study in a Tertiary-Care Hospital in Hungary. Diseases 2019, 7, e41.
- Mulchany, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2014, 68, 1–12.
- Ciszek-Lenda, M.; Strus, M.; Walczewska, M.; Majka, G.; Machul-Żwirbla, A.; Mikolajczyk, D.; Górska, S.; Gamian, A.; Chain, B.; Marcinkiewicz, J. Pseudomonas aeruginosa biofilm is a potent inducer of phagocyte hyperinflammation. Inflamm. Res. 2019, 68, 397–413.
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, e913.
- Chen, H.; Wubbolts, R.W.; Haagsman, H.P.; Weldhuizen, E.J.A. Inhibition and Eradication of Pseudomonas aeruginosa Biofilms by Host Defence Peptides. Sci. Rep. 2018, 8, e10446.
- Barrak, I.; Baráth, Z.; Tián, T.; Venkei, A.; Gajdács, M.; Urbán, E.; Stájer, A. Effects of different decontaminating solutions used for the treatment of peri-implantitis on the growth of Porphyromonas gingivalis—An in vitro study. Acta Microbiol. Immunol. Hung. 2020, doi:10.1556/030.2020.01176.
- Stájer, A.; Barrak, I.; Gajdács, M.; Urbán, E.; Baráth, Z. Diagnosis and Management of Cervicofacial Actinomycosis: Les-sons from Two Distinct Clinical Cases. Antibiotics 2020, 9, e139.
- Thuenauer, R.; Landi, A.; Trefzer, A.; Altmann, S.; Wehrum, S.; Elerhoff, T.; Diedrich, B.; Dengjel, J.; Nyström, A.; Imberty, A.; et al. The Pseudomonas aeruginosa Lectin LecB Causes Integrin Internalization and Inhibits Epithelial Wound Healing. mBio 2020, 11, e03260-19.
- Brown, V.I.; Lowbury, E.J.L. Use of an improved cetrimide agar medium and other culture methods for Pseudomonas aeruginosa. J. Clin. Pathol. 1965, 18, 752–756.
- Martínez-Alemán, S.; Bustamante, A.E.; Jimenez-Valdes, R.J.; González, G.M.; Sánchez-González, A. Pseudomonas aeruginosa isolates from cystic fibrosis patients induce neutrophil extracellular traps with different morphologies that could correlate with their disease severity. Int. J. Med. Microbiol. 2020, 310, e151451.
- Martínez-Alemán, S.; Bustamante, A.E.; Jimenez-Valdes, R.J.; González, G.M.; Sánchez-González, A. Pseudomonas aeru-ginosa isolates from cystic fibrosis patients induce neutrophil extracellular traps with different morphologies that could correlate with their disease severity. Int. J. Med. Microbiol. 2020, 310, e151451.
- Schinner, S.; Engelhardt, F.; Preusse, M.; Thöming, J.G.; Tomasch, J.; Haussler, S. Genetic determinants of Pseudomonas aeruginosa fitness during biofilm growth. Biofilm 2020, 1, e100023.
- Trunk, K.; Benkert, B.; Quack, N.; Münch, R.; Scheer, M.; Garbe, J.; Jansch, L.; Trost, M.; Wehland, J.; Buer, J.; et al. Anaerobic adaptation in Pseudomonas aeruginosa: Definition of the Anr and Dnr regulons. Environ. Microbiol. 2010, 12, 1719–1733
- Hoiby, N.; Ciofu, O.; Bjarnsholt, T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010, 5, 1663–1674.
- Grassi, L.; Di Luca, M.; Maisetta, G.; Rinaldi, A.C.; Esin, S.; Trampuz, A.; Batoni, G. Generation of Persister Cells of Pseu-domonas aeruginosa and Staphylococcus aureus by Chemical Treatment and Evaluation of Their Susceptibility to Mem-brane-Targeting Agents. Front. Microbiol. 2017, 8, e1917.
- Balogh, E.P.; Faludi, I.; Virók, D.P.; Endrész, V.; Burián, K. Chlamydophila pneumoniae induces production of the defen-sin-like MIG/CXCL9, which has in vitro antichlamydial activity. Int. J. Med. Microbiol. 2011, 301, 252–259.
- Burián, K.; Berencsi, K.; Endrész, V.; Gyulai, Z.; Vályi-Nagy, T.; Bakay, M.; Geng, Y.; Virók, D.; Kari, L.; Hajnal-Papp, R.; et al. Chlamydia pneumoniae Exacerbates Aortic Inflammatory Foci Caused by Murine Cytomegalovirus Infection in Normocholesterolemic Mice. Clin. Vaccine Immunol. 2001, 8, 1263–1266.
- Möker, N.; Dean, C.R.; Tao, J. Pseudomonas aeruginosaIncreases Formation of Multidrug-TolerantPersister Cells in Re-sponse to Quorum-Sensing Signaling Molecule. J. Bacteriol. 2010, 192, 1946–1955.
- Li, M.; Long, Y.; Liu, Y.; Liu, Y.; Chen, R.; Zhang, L.; Jin, Y.; Yang, L.; Bai, F.; Jin, S.; et al. HigB of Pseudomonas aeruginosa Enhances Killing of Phagocytes by Up-Regulating the Type III Secretion System in Ciprofloxacin Induced Persister Cells. Front. Cell. Infect. Microbiol. 2016, 6, e125.
- Gnanadhas, D.P.; Elango, M.; Datey, A.; Chakravortty, D. Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by L-Methionine in combination with antibiotic therapy. Sci. Rep. 2015, 5, e16043.
- Alenanizy, F.S.; Alqahtani, F.Y.; Eltayb, E.K.; Alrumikan, N.; Almebki, R.; Alhossan, A.; Almangour, T.A.; Al-Qahtani, H. Evaluating the effect of antibiotics sub-inhibitory dose on Pseudomonas aeruginosa quorum sensing dependent viru-lence and its phenotypes. Saudi J. Biol. Sci. 2020, doi:10.1016/j.sjbs.2020.10.040.
- Hemati, S.; Kouhsari, E.; Sadeghifard, N.; Maleki, A.; Omidi, N.; Mahdavi, Z.; Pakzad, I. Sub-minimum inhibitory con-centrations of biocides induced biofilm formation in Pseudomonas aeruginosa. New Microbe New Infect. 2020, 38, e100794.
- Navidifar, T.; Amin, M.; Rashno, M. Effects of sub-inhibitory concentrations of meropenem and tigecycline on the expres-sion of genes regulating pili, efflux pumps and virulence factors involved in biofilm formation by Acinetobacter bau-mannii. Infect. Drug Resist. 2019, 12, 1099–1111.
- Subedi, D.; Vijay, A.K.; Wilcox, M. Study of Disinfectant Resistance Genes in Ocular Isolates of Pseudomonas aeruginosa. Antibiotics 2018, 7, e88.
- Zhu, L.; Lin, J.; Ma, J.; Cronan, J.E.; Wang, H. Triclosan Resistance of Pseudomonas aeruginosa PAO1 Is Due to FabV, a Triclosan-Resistant Enoyl-Acyl Carrier Protein Reductase. Antimicrob. Agents Chemother. 2010, 54, 689–698.
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Minervini, F.; Limitone, A. Cell–cell communication in food related bacteria. Int. J. Food Microbiol. 2007, 120, 34–45.
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199.
- Pesci, E.C.; Iglewski, B.H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997, 2, 132–134.
- Turan, N.B.; Chormey, D.S.; Büyükpinar, C.; Engin, G.O.; Bakirdere, S. Quorum sensing: Little talks for an effective bac-terial coordination. Trends Anal. Chem. 2017, 91, 1–11.
- Rampioni, G.; Leoni, L.; Williams, P. The art of antibacterial warfare: Deception through interference with quorum sens-ing–mediated communication. Bioorg. Chem. 2014, 55, 60–68.
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Minervini, F.; Limitone, A. Cell–cell communication in food related bacteria. Int. J. Food Microbiol. 2007, 120, 34–45.
- Fuse, K.; Fujimura, S.; Kikuchi, T.; Gomi, K.; Iida, Y.; Nukiwa, T.; Watanabe, A. Reduction of virulence factor pyocyanin production in multidrug-resistant Pseudomonas aeruginosa. J. Infect. Chemother. 2013, 19, 82–88.
- Whiteley, M.; Lee, K.M.; Greenberg, E. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 13904–13909.
- Venturi, V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006, 30, 274–291.
- Smith, R.S.; Iglewski, B.H. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 2003, 112, 1460–1465.
- Duan, K.; Surette, M.G.; Environmental Regulation of Pseudomonas aeruginosa PAO1 Las and Rhl Quorum-Sensing Systems. J. Bacteriol. 2007, 189, 4827–4836.
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41.
- 114. Postat, J.; Bousso, P. Quorum Sensing by Monocyte-Derived Populations. Front. Immunol. 2019, 10, e2140.
- Shih, P.C.; Huang, C.T. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibi-otic resistance. J. Antimicrob. Chemother. 2002, 49, 309–314.
- Geisinger, E.; Isberg, R.R. Interplay between Antibiotic Resistance and Virulence During Disease Promoted by Multi-drug-Resistant Bacteria. J. Infect. Dis. 2017, 215, S9–S17.
- Rumbaugh, K.P.; Griswold, J.A.; Iglewski, B.H.; Hamood, A.N. Contribution of Quorum Sensing to the Virulence of Pseudomonas aeruginosa in Burn Wound Infections. Infect Immun. 1999, 67, 5854–5862.
- Krzyzek, P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front. Microbiol. 2019, 10, e2473.
- Rada, B.; Leto, T.L. Pyocyanin effects on respiratory epithelium: Relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013, 21, 73–81.
- Song, F.; Wang, H.; Sauer, K.; Ren, D. Cyclic-di-GMP and oprF Are Involved in the Response of Pseudomonas aeruginosa to Substrate Material Stiffness during Attachment on Polydimethylsiloxane (PDMS). Front. Microbiol. 2018, 9, 110.
- Valentini, M.; Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555.