
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ROESELERS Guus | + 2968 word(s) | 2968 | 2020-12-30 12:17:27 | | | |
| 2 | ROESELERS Guus | -96 word(s) | 2872 | 2021-01-06 12:47:56 | | | | |
| 3 | Peter Tang | -5 word(s) | 2867 | 2021-01-09 15:04:57 | | |
Video Upload Options
There are several parallel mechanisms by which early life microbiome acquisition may proceed, including early exposure to maternal vaginal and fecal microbiota, transmission of skin associated microbes, and ingestion of microorganisms present in breast milk. The crucial role of vertical transmission from the maternal microbial reservoir during vaginal delivery is supported by the shared microbial strains observed among mothers and their babies and the distinctly different gut microbiome composition of caesarean-section born infants. The healthy infant colon is often dominated by members of the keystone genus Bifidobacterium that have evolved complex genetic pathways to metabolize different glycans present in human milk. In exchange for these host-derived nutrients, bifidobacteria's saccharolytic activity results in an anaerobic and acidic gut environment that is protective against enteropathogenic infection. Interference with early-life microbiota acquisition and development could result in adverse health outcomes. Compromised microbiota development, often characterized by decreased abundance of Bifidobacterium species has been reported in infants delivered prematurely, delivered by caesarean section, early life antibiotic exposure and in the case of early life allergies. Various microbiome modulation strategies such as probiotic, prebiotics, synbiotics and postbiotics have been developed that are able to generate a bifidogenic shift and help to restore the microbiota development.
1. Introduction
Intestinal microbiota development in early life is very dynamic and is in synergy with anatomical, intestinal physiology, immune and neurological development [1]. Deviations and misconfigurations in its structure and function may contribute to pathologies and chronic diseased states [2]. Timing of the first microbial exposure to the developing embryo is often debated [3]: some reports suggest that microbial exposure and colonization already happens "in utero" [4][5][6] while others have pointed out that more detailed studies are required to verify this observation [3].
Undoubtedly, there are many more parsimonious routes by which microbial inoculation may proceed, including early exposure to vaginal and fecal microbiota at birth, ingestion of epidermal skin microbes and viable microbes present in breast milk, as well as the inherently close interactions between the neonate and mother [7]. The subsequent colonization process is a balance between influx of microbes and niche adaptability [8].
These first microbial pioneers become key players in the assembly of a complex ecosystem that follows distinct successional stages with potential long term health consequences [9]. The formation of this complex ecosystem is influenced by multiple factors including host genetics, mother's microbiota, gestational age, medical practices, mode of delivery, diet, life style, familial environment, presence of pets, infectious diseases and antimicrobial therapies [10]. Although microbiota assembly in the infant gut is not strictly deterministic, there is an overarching directionality of microbial succession strongly driven by early life nutrition, specifically human breast milk.
Human milk not only provides optimal nutrition for the infants but is also a reservoir of microbes mainly within the genera Staphylococcus and Streptococcus but also Bifidobacterium, Propionibacteria, Pseudomonas, Bacteroides and Parabacteroides [11][12]. Bifidobacterium is the most abundant genus in the breastfed infant's gut and is considered a true "keystone" taxon with a strong eco-physiological impact on microbiota composition and activity. Therefore, Bifidobacterium spp. may serve as a marker of healthy microbiota development and breast-feeding practices.
2. Ecological Drivers of Acquisition and Succession of Bifidobacteria
2.1. Mode of Delivery, Antibiotics and Diet
Immediately after birth, the infant gut is still rich in oxygen and offers a favorable habitat for facultative anaerobic microorganisms such as Staphylococcus spp., Streptococcus spp., Enterobacter spp. and other members of family Enterobacteriaceae [10]. These pioneering species play an important role in the rapid transition from a microbiome dominated by taxa that tolerate or thrive under limited oxygen to microbiome dominated by strictly anaerobic taxa such as Clostridium, Bacteroides, Eubacterium and Bifidobacterium spp. [2][10]. However, birth via caesarean section (c-section) interrupts this program of microbiota acquisition and colonization since there is no contact with the maternal vaginal and fecal microbiota and the perineal skin. In addition, c-section born infants are often exposed to maternal prophylactic antibiotic administration and c-section birth has been shown to adversely affect breastfeeding initiation, milk supply and infant breastfeeding receptivity compared to vaginal deliveries, which may further compromise early life microbiota development. [13].
Instead, the guts of c-section-delivered infants are typically first colonized by human skin and oral cavity associated bacteria, which include Staphylococcus spp., Streptococcus spp., Veillonella spp., Propionibacterium spp., Corynebacterium spp. and Acinetobacter spp. [7][14][15]. Recently, a large study with 596 infants confirmed microbiota differences observed in c-section born infants [16]. Notably, this study reported that in vaginal born infants, commensal genera, such as Bacteroides and Bifidobacterium (such as Bifidobacterium longum and Bifidobacterium breve and Bifidobacterium infantis), made up 68% of the total genus richness, while c-section born infants were depleted of these commensal genera and instead were enriched by species within the genera Enterococcus, Staphylococcus, Streptococcus, Klebsiella, Enterobacter and Clostridium, all of which are more characteristic for hospital environments and hospitalized preterm babies [16]. In addition, c-section delivery is often accompanied with varying use of medications, including prophylactic antibiotics, which further adds to the disrupted transfer of maternal gut microbiota.
It has been shown that direct administration of antibiotics to neonates or indirect through feto-placental circulation has pervasive effects on gut microbiota composition, and is associated with adverse immune outcomes such as allergies and atopies [17], and metabolic health outcomes such as obesity [18]. Fouhy et al. demonstrated that antibiotic administration to term neonates led to a relative increase in fecal Proteobacteria and a decrease in Actinobacteria, particularly Bifidobacterium spp., representing deviation from normal microbiota development [19]. Intriguingly, it was also shown that intrapartum antibiotic administration not only led to differences in infant gut microbiota but also affected breast milk microbiota composition [20].
Upon delivery, breast milk is the most significant factor which impacts maturation of the gut microbiota. It has been shown that formula-fed infants exhibit a more diverse microbiota than breastfed infants [9][21][22]. The microbiota of vaginally delivered breast-fed infants is dominated by the Phylum Actinobacteria, while formula-fed infants adopt a more diverse microbiota [23]. Moreover, the cessation of breast feeding is associated with a steep reduction in the levels of bifidobacteria and an accelerated increase in members of Firmicutes and Bacteroides, which further substantiates the key role of the milk–bifidobacteria interaction in shaping the human gut microbiota [9].
2.2. Establishment of Bifidobacterium: A Keystone Genus
Bifidobacteria are regarded as a "keystone" taxon in the early life gut microbiota. The keystone organism concept is a fundamental principle in theoretical and applied ecology. The term was first introduced in a study on a rocky intertidal ecosystem in California. When the top predator (a starfish) was removed, the community collapsed, prompting the architectural analogy with the keystone of an arch [24]. Keystone organisms are important drivers of community structure and integrity, and their influence is non-redundant. Their activity and biotic interactions disproportionately determine species assemblages and the rates of material and energy flow across entire communities. These taxa have a unique and crucial role in microbial communities, and their removal can cause a dramatic shift in microbiome structure and functioning.
By their broad impact on microbiota composition function, keystone members are also likely to exert strong direct and indirect effects on host physiology and may be essential for host homeostasis and health [25]. In early life gut microbiota, Bifidobacterium and Bacteroides drive microbiota development by maintaining a strict anaerobic environment, by producing and cross-feeding on metabolites such as short chain fatty acids (SCFA). SCFA result in a low pH, which is among the main mechanisms of ecological resistance against pathogens [26][27]. The failure of these keystone taxa to colonize and drive succession may lead or contribute to the development of chronic diseases [28]. For instance, antibiotics administration has been clearly demonstrated to impair proliferation of these keystone taxa, which could in turn disturb microbial interaction with the immune system, particularly during critical stages of development. This failure of cross talk between keystone microbes and immune cells is thought to be an important factor in the development of allergies, metabolic disorders and infectious diseases [29].
Physiologically, the keystone function of bifidobacteria is strongly linked to their unique metabolic capacity and genomic architecture. Bifidobacteria are genetically adapted to utilize specific glycans of human milk, thus representing an intriguing example of host-microbe coevolution into mutualistic symbiosis.
3. Evolutionary and Eco-Physiological Attributes of a Bifidogenic Milieu
3.1. Evolutionary Ecology
The heterofermentative genus Bifidobacterium takes its name from its characteristic Y-shaped cells (in Latin, bifidus means cleft or divided into two parts). Isolation of bifidobacteria from infant feces provided first indications of their ecological relevance in the human gut [30]. Interestingly, bifidobacteria were also isolated from the gut of multicellular social organisms such as birds, mammals and social insects, which implies that bifidobacteria could be transmitted vertically [31][32]. In the gut, growth of bifidobacteria is nurtured by glycans through feeding or cross-feeding activity [33]. The unique genetic make-up of Bifidobacterium species gives an advantage to outcompete other gut commensals in metabolizing glycans present in human milk [30]. These two exceptional features—maternal transfer and genetic constitution—distinguish bifidobacteria from all other commensal gut bacteria such as Lactobacillus species.
Several studies investigating specific maternal–neonatal microbial transference have shown that mother and child often share genomically identical bifidobacterial strains belonging to B. breve and B. longum subsp. longum, B. longum subsp. Infantis, further substantiating vertical transmission of bifidobacteria (Figure 1) [34][35][36][37]. These findings provide initial insights as to why vaginal delivery provides a higher abundance of Bifidobacterium species in infants, over a c-section delivery.
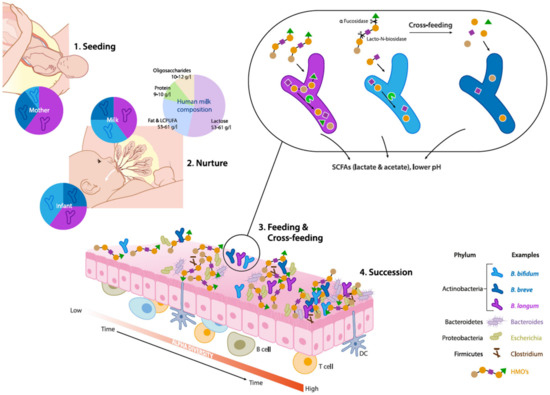
Figure 1. Schematic overview of eco-physiological factors driving maternal transmission, colonization and succession of Bifidobacterium species in the early life gut microbiota. (1) Seeding: Bifidobacteria are transmitted from mother to child during vaginal delivery. (2) Nurture: Human milk contains viable microorganisms, including Bifidobacterium species, which contribute to seeding the infant's gut microbiota. Human milk oligosaccharides (HMOs) are minimally digested by the infant and metabolized by infant-type bifidobacteria by convergent mechanisms. (3) Feeding & Cross-feeding: different Bifidobacterium species and strains have distinct HMO degrading abilities resulting in varied HMO consumption behaviors. Degradation of HMO occurs sequentially with the removal monosaccharides and requires a multitude of enzymes with various glycosidic specificities. Bifidobacterial cross-feeding permits the sharing of resources to maximize nutrient consumption from the diet and highlights the cooperative nature of bifidobacterial strains and their role as "key stone" species in the infant gut microbiota. (4) Succession: The combined activity of Bifidobacterium species contributes to the establishment and maintenance of a strict anaerobic environment and low pH by producing metabolites such as lactate and acetate. These conditions allow the successive establishment of butyrate producing taxa such as Eubacterium and Anaerostipes spp which are characteristic for a more matured microbiota.
In vaginally delivered breast-fed infants, the relative abundance of bifidobacteria could be over 90% but typically decreases to less than 5% in adults [38][39]. More specifically, breast-fed infants are predominated by the presence of B. breve spp., B. bifidum, B. longum subsp. infantis and B. longum subsp. longum, also known as infant type Human Resident Bifidobacteria (HRB) (Figure 1) [32]. On the other hand, adults are characterized by the presence of B. adolescentis and B. catenulatum, B. pseudocatenulatum and B. longum subsp. longum which are often termed as adult-type HRB [32][33]. Notably, B. longum subsp. longum was found to be predominant in both infant and adult gut. Hence, there is no strict distinction between "infant" type and "adult" type bifidobacteria, as some of the adult bifidobacteria such as B. adolescentis were shown to be vertically transferred to infants [40].
3.2. Genomic Features
The typical characteristics of bifidobacteria include their obligate anaerobism, peptidoglycan rich cell walls and heterofermentative metabolism. At present, there are 51 species and 10 subspecies of bifidobacteria reported, out of which 48 (sub)species have at least one genome sequenced (NCBI database) [41]. Pan-genome analyses of the Bifidobacterium genus revealed that 13.7% of the identified bifidobacterial genes are involved in carbohydrate metabolism, which is much higher than the other analyzed gut commensals [30][42]. Importantly, the core genome also encodes enzymes involved in the "bifid shunt", which equips bifidobacteria with a unique evolutionary advantage of generating more ATP (per mole of glucose) in comparison to microorganisms using other carbohydrate fermentative pathways such as glycolysis [30]. Specifically, a Cluster of Orthologous Groups (COGs) representing α-amylases, ATP-binding cassettes (ABC) and phosphoenolpyruvate-phosphotransferase systems (PEP-PTS) were identified to be acquired during the course of evolution and give bifidobacteria a selective advantage in the highly competitive ecological niche of the early life gut [42]. Furthermore, comparative genome analyses of bifidobacteria, particularly B. longum subsp. infantis ATCC15697 and B. bifidum PRL2010, have revealed that these species are able to utilize a broad range of host-derived glycans (HMOs and mucin), further corroborating the genomic plasticity of Bifidobacterium spp. [30].
3.3. HMO Utilization
Human milk constitutes around 10–12 gm/L of oligosaccharides, which constitute the third most abundant component in milk [43][44] (Figure 1). HMOs are complex and structurally highly diverse, with over 200 different molecules that vary in size, Degree of Polymerization (DP), charge and sequence. The size distribution of HMOs ranges from 90% short chain oligosaccharides to 10% long chain oligosaccharides [45]. The most basic HMO structures are monomers of glucose (Glc), galactose (Gal) and N-acetylglucosamine (GlcNAc), and also contain fucose (Fuc) and/or N-acetylneuraminic acid (NeuAc) linked via several glycosidic bonds [46]. Most HMOs cannot be utilized by host digestive enzymes but are effectively utilized by gut microbiota. Therefore, HMOs play a pivotal role in shaping the infant gut microbiota, and actively promote beneficial bacteria, which is also termed as a "prebiotic" effect [47].
The abundance and prevalence of Bifidobacteria in the neonatal gut is attributed to their unique ability to catabolize HMOs [48]. For example, B. longum subsp. infantis and B. breve use specific ATP-binding cassette (ABC) transporters for internalization of intact oligosaccharides. Intracellular glycosyl hydrolases (GH) such as fucosidases, hexosaminidases and sialidases can further deconstruct the oligosaccharides [49][50] (Figure 1). Species such as B. bifidum have different HMO consumption capabilities. These taxa break down HMO via extracellular glycosidases into mono- and disaccharides, which are subsequently transported into the cells via permeases. Residues of this extracellular degradation allow cross-feeding of other types of bacteria including other Bifidobacterium species [51].
Preclinical experiments in conventional mice receiving a combination B. bifidum PRL2010, B. longum subsp. infantis ATCC15697, B. adolescentis 22L, and B. breve 12L, demonstrated a synergistic effect by acting directly or by cross-feeding on host or plant derived carbohydrates, which further led to the enrichment of murine gut glycobiome [52]. In contrast, other gut commensals such as Lactobacillus sp. and Bacteroides sp. show poor or limited capacity to utilize HMOs, respectively [50][53].
Short chain galacto-oligosaccharides (scGOS) and long chain fructo-oligosaccharides (lcFOS) in a ratio of 9:1 mimic the size distribution of HMOs and resembles functionality of breast milk [54]. Based on these observations, scGOS/lcFOS, and synthetic HMOs such as 2′Fucosyl Lactose (2′FL) and Lacto-N-neotetraose (LNnT), are being incorporated in infant formula. Altogether, these chemical constituents or prebiotics are aimed to increase bifidobacteria counts and thereby exert immune benefits. In clinical studies, it was demonstrated that 2′FL and LNnT modulated the gut microbiota exhibiting increased levels of Actinobacteria, specifically Bifidobacterium spp., and decreased levels of Firmicutes and Proteobacteria [55][56].
3.4. Effect on pH and SCFA Production
In breast-fed infants, HMO metabolism by gut microbiota is often associated with distinct SCFA profiles when compared to formula-fed infants, which are subsequently reflected in reduced fecal pH [57][58]. The prevalent SCFAs include acetate, butyrate, valerate, propionate and, to a lesser extent, branched chain fatty acids such as iso-butyrate and iso-valerate. Exclusively breast-fed infants are characterized by a higher relative proportion of acetate relative to other SCFAs in the gut, which was found to be independent of birth mode, sex, intrapartum antibiotics, site of recruitment and maternal body mass index [57][58][59]. Importantly, Bifidobacteriaceae was the only family which was significantly associated with fecal pH, although there are microbes (such as Bacteroidaceae) which can also utilize HMO [27][60]. In particular, bifidobacteria have evolved with specific mechanisms to produce acetic and lactic acids (in a molar ratio of 3:2) by utilization of glycans through the bifid shunt pathway (Figure 1). [61]. Based on these observations, fecal acetate is also regarded as a biomarker for bifidogenic activity and overall microbiota health in early life.
Physiologically, SCFAs have also been associated with both systemic effects such as immune modulation and local effects such as acting as an energy source for colonocytes. SCFAs may also provide colonization resistance against pathogens such as Escherichia coli O157:H7 [57][62]. In a clinical study, reduced fecal acetate at 3 months of age was associated with atopic wheeze observed 9 months later [63]. In addition, it was recently shown that prebiotic supplementation (short chain galacto-oligosaccharides (scGOS) and long-chain fructo-oligosaccharides (lcFOS)) in a partially hydrolyzed protein formula, led to increased Bifidobacterium/Lachnospiraceae ratio, which in turn was reflected in organic acid profiles with high acetate and lactate levels and low butyrate, propionate and branched chain SCFAs [64].
On the other hand, butyrate is considered as a marker for a healthy maturation of the gut microbiota when the infant diet diversifies with the introduction of solid foods [58][65]. Interestingly, acetate and lactate are important "cross-feeding" substrates for butyrate-producing bacteria such as Faecalibacterium prausnitzii, Roseburia, Anaerostipes spp. and Eubacterium halli (Figure 1) [66]. This gradual transition from a bifidogenic and acetogenic milieu towards a butyrogenic milieu (more adult like) may be of critical importance for a healthy maturation of the gut and the gut microbiota. Wopereis et al. (2017) proposed that this maturation process is associated with a reduced risk of developing eczema in infants at risk of developing allergies [64]. Moreover, butyrate has been shown to have regulatory effects on host immunity including anti-inflammatory mechanisms and has been generally associated with numerous health benefits by improvement of gut barrier function and pathogen inhibition [67].
References
- Wopereis: H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014, 25, 428–438, doi:10.1111/pai.12232.
- Houghteling, P.D.; Walker, W.A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307, doi:10.1097/MPG.0000000000000597.
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48, doi:10.1186/s40168-017-0268-4.
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, doi:10.1126/scitranslmed.3008599.
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, doi:10.1016/j.resmic.2007.12.007.
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, doi:10.1038/srep23129.
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, doi:10.1073/pnas.1002601107.
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133-145.e135, doi:10.1016/j.chom.2018.06.005.
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588, doi:10.1038/s41586-018-0617-x.
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173, doi:10.1016/j.tim.2012.12.001.
- Makino, H.; Martin, R.; Ishikawa, E.; Gawad, A.; Kubota, H.; Sakai, T.; Oishi, K.; Tanaka, R.; Ben-Amor, K.; Knol, J.; et al. Multilocus sequence typing of bifidobacterial strains from infant’s faeces and human milk: Are bifidobacteria being sustainably shared during breastfeeding? Benefic. Microbes 2015, 6, 563–572, doi:10.3920/BM2014.0082.
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C., et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 2016, 7, doi:10.3389/fmicb.2016.01619.
- Hobbs, A.J.; Mannion, C.A.; McDonald, S.W.; Brockway, M.; Tough, S.C. The impact of caesarean section on breastfeeding initiation, duration and difficulties in the first four months postpartum. BMC Pregnancy Childbirth 2016, 16, 90, doi:10.1186/s12884-016-0876-1.
- Scholtens, P.A.M.J.; Oozeer, R.; Martin, R.; Amor, K.B.; Knol, J. The early settlers: Intestinal microbiology in early life. Annu. Rev. Food Sci. Technol. 2012, 3, 425–447, doi:10.1146/annurev-food-022811-101120.
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703, doi:10.1016/j.chom.2015.04.004.
- Yan, S. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, doi:10.1038/s41586-019-1560-1.
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Lang, J.E.; Garssen, J.; Kraneveld, A.; Maitland-van der Zee, A.H. Early-life antibiotic exposure increases the risk of developing allergic symptoms later in life: A meta-analysis. Allergy 2018, 73, 971–986, doi:10.1111/all.13332.
- Turta, O.; Rautava, S. Antibiotics, obesity and the link to microbes—What are we doing to our children? BMC Med. 2016, 14, 57, doi:10.1186/s12916-016-0605-7.
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820, doi:10.1128/AAC.00789-12.
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 2019, 6, doi:10.3389/fnut.2019.00004.
- di Gioia, D.; Aloisio, I.; Mazzola, G.; Biavati, B. Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl. Microbiol. Biotechnol. 2014, 98, 563–577, doi:10.1007/s00253-013-5405-9.
- Martin, R.; Makino, H.; Yavuz, A.C.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS ONE 2016, 11, doi:10.1371/journal.pone.0158498.
- Voreades, N.; Kozil, A.; Weir, T.L. Diet and the development of the human intestinal microbiome. Front. Microbiol. 2014, 5, doi:10.3389/fmicb.2014.00494.
- Paine, R.T. A note on trophic complexity and community stability. Am. Nat. 1969, 103, 91–93, doi:10.1086/282586.
- Newburg, D.S.; Morelli, L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr. Res. 2015, 77, 115–120, doi:10.1038/pr.2014.178.
- Laforest-Lapointe, I.; Arrieta, M.-C. Patterns of early-life gut microbial colonization during human immune development: An ecological perspective. Front. Immunol. 2017, 8, 788, doi:10.3389/fimmu.2017.00788.
- Henrick, B.; Hutton, A.; Palumbo, M.; Casaburi, G.; Smilowitz, J.; Underwood, M.; Frese, S. Changes in infant fecal pH shows a population-wide, generational loss of Bifidobacterium among breastfed infants over the last century. J. Pediatric. Gastroenterol. Nutr. 2018, 66, 1073.
- Yu, Z.-T.; Chen, C.; Kling, D.E.; Liu, B.; McCoy, J.M.; Merighi, M.; Heidtman, M.; Newburg, D.S. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology 2013, 23, 169–177, doi:10.1093/glycob/cws138.
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564, doi:10.1016/j.chom.2015.04.006.
- Milani, C.; Turroni, F.; Duranti, S.; Lugli, G.A.; Mancabelli, L.; Ferrario, C.; van Sinderen, D.; Ventura, M. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl .Environ. Microbiol. 2015, 82, 980–991, doi:10.1128/AEM.03500-15.
- Ventura, M.; Turroni, F.; Lugli, G.A.; van Sinderen, D. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J. Sci. Food Agric. 2014, 94, 163–168, doi:10.1002/jsfa.6356.
- Wong, C.B.; Sugahara, H.; Odamaki, T.; Xiao, J.Z. Different physiological properties of human-residential and non-human-residential bifidobacteria in human health. Benefic. Microbes 2017, 9, 111–122, doi:10.3920/BM2017.0031.
- Turroni, F.; Milani, C.; Duranti, S.; Mahony, J.; van Sinderen, D.; Ventura, M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2017, doi:10.1016/j.tim.2017.10.001.
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015, 81, 7078–7087, doi:10.1128/aem.02037-15.
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Ben Amor, K.; Oozeer, R.; et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793, doi:10.1128/AEM.05346-11.
- Korpela, K.; Costea, P.; Coelho, L.P.; Kandels-Lewis, S.; Willemsen, G.; Boomsma, D.I.; Segata, N.; Bork, P. Selective maternal seeding and environment shape the human gut microbiome. Genome. Res. 2018, 28, 561–568, doi:10.1101/gr.233940.117.
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66, doi:10.1186/s40168-017-0282-6.
- Vaiserman, A.M.; Koliada, A.K.; Marotta, F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 2017, 35, 36–45, doi:10.1016/j.arr.2017.01.001.
- Kato, K.; Odamaki, T.; Mitsuyama, E.; Sugahara, H.; Xiao, J.-z.; Osawa, R. Age-related changes in the composition of gut bifidobacterium species. Curr. Microbiol. 2017, 74, 987–995, doi:10.1007/s00284-017-1272-4.
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204, doi:10.3389/fmicb.2016.01204.
- Bottacini, F.; van Sinderen, D.; Ventura, M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem. J. 2017, 474, 4137–4152, doi:10.1042/BCJ20160756.
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Bottacini, F.; Mangifesta, M.; Sanchez, B.; Viappiani, A.; Mancabelli, L.; Taminiau, B.; et al. Genomic encyclopedia of type strains of the genus bifidobacterium. Appl. Environ. Microbiol. 2014, 80, 6290–6302, doi:10.1128/aem.02308-14.
- Meurant, G. Handbook of Milk Composition; Elsevier Science: Amsterdam, The Netherlands, 1995.
- Zivkovic, A.M.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 4653.
- Stahl, B.; Thurl, S.; Zeng, J.; Karas, M.; Hillenkamp, F.; Steup, M.; Sawatzki, G. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 1994, 223, 218–226, doi:10.1006/abio.1994.1577.
- Ruiz-Moyano, S.; Totten, S.M.; Garrido, D.A.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl. Environ. Microbiol. 2013, 79, 6040–6049, doi:10.1128/AEM.01843-13.
- Thomson, P.; Medina, D.A.; Garrido, D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018, 75, 37–46, doi:10.1016/j.fm.2017.09.001.
- Sakanaka, M.; Hansen, M.E.; Gotoh, A.; Katoh, T.; Yoshida, K.; Odamaki, T.; Yachi, H.; Sugiyama, Y.; Kurihara, S.; Hirose, J.; et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019, 5, eaaw7696, doi:10.1126/sciadv.aaw7696 %J Science Advances.
- Garrido, D.; Barile, D.; Mills, D.A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012, 3, 415S–421S, doi:10.3945/an.111.001586.
- Zúñiga, M.; Monedero, V.; Yebra, M.J. Utilization of host-derived glycans by intestinal lactobacillus and bifidobacterium species. Front. Microbiol. 2018, 9, doi:10.3389/fmicb.2018.01917.
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782, doi:10.1038/srep15782
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. Isme J. 2016, 10, 1656, doi:10.1038/ismej.2015.236
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr. Rev. 2015, 73, 426–437, doi:10.1093/nutrit/nuu016.
- Boehm, G.; Moro, G. Structural and functional aspects of prebiotics used in infant nutrition. J. Nutr. Suppl. 2008, 138, 1818s–1828s.
- Alliet, P.; Puccio, G.; Janssens, E.; Cajozzo, C.; Corsello, G.; Berger, B.; Sperisen, P.; Martin, F.-P.; Sprenger, N.; Steenhout, P. Term Infant Formula Supplemented with Human Milk Oligosaccharides (2′fucosyllactose and Lacto-Neotetraose) Shifts Stoom Microbiota and Metabolic Signatures Closer to that of Breastfed Infants. J. Pediatric Gastroenterol. Nutr. 2016, 63, S55, doi:10.1097/01.mpg.0000489632.17881.57.
- Elison, E.; Vigsnaes, L.K.; Rindom Krogsgaard, L.; Rasmussen, J.; Sørensen, N.; McConnell, B.; Hennet, T.; Sommer, M.O.A.; Bytzer, P. Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br. J. Nutr. 2016, 116, 1356–1368, doi:10.1017/S0007114516003354.
- Bridgman, S.L.; Azad, M.B.; Field, C.J.; Haqq, A.M.; Becker, A.B.; Mandhane, P.J.; Subbarao, P.; Turvey, S.E.; Sears, M.R.; Scott, J.A.; et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: Differences in relative versus absolute concentrations. Front. Nutr. 2017, 4, doi:10.3389/fnut.2017.00011.
- Pham, V.T.; Lacroix, C.; Braegger, C.P.; Chassard, C. Early colonization of functional groups of microbes in the infant gut. Environ. Microbiol. 2016, 18, 2246–2258, doi:10.1111/1462-2920.13316.
- Edwards, C.A.; Parrett, A.M.; Balmer, S.E.; Wharton, B.A. Faecal short chain fatty acids in breast-fed and formula-fed babies. Acta Paediatr. 1994, 83, 459–462, doi:doi:10.1111/j.1651-2227.1994.tb13059.x.
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century mSphere 2018, 3, e00041-00018, doi:10.1128/mSphere.00041-18.
- Orrhage, K.; Nord, C.E. Bifidobacteria and lactobacilli in human health. Drugs Exp. Clin. Res. 2000, 26, 95–111.
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547, doi:10.1038/nature09646.
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152, doi:10.1126/scitranslmed.aab2271.
- Wopereis, H.; Sim, K.; Shaw, A.; Warner, J.O.; Knol, J.; Kroll, J.S. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J. Allergy Clin. Immunol. 2017, 10.1016/j.jaci.2017.05.054, doi:10.1016/j.jaci.2017.05.054.
- Kumari, M.; Kozyrskyj, A.L. Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obes. Rev. 2017, 18, 18–31, doi:doi:10.1111/obr.12484.
- Schwab, C.; Ruscheweyh, H.-J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic Interactions of infant bifidobacteria and eubacterium hallii during L-Fucose and fucosyllactose degradation. Front. Microbiol. 2017, 8, doi:10.3389/fmicb.2017.00095.
- Lee, H.; Shen, H.; Hwang, I.; Ling, H.; Yew, W.; Lee, Y.; Chang, M. Targeted Approaches for in situ gut microbiome manipulation. Genes 2018, 9, 351.




