| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexandre Trindade | + 1712 word(s) | 1712 | 2020-11-19 08:57:41 |
Video Upload Options
The Notch signaling pathway is a major regulator of vascular morphogenesis, managing endothelial response to vascular growth factors, endothelial specialization, establishment and maintenance of vascular identity as venous or arterial and vascular maturation.
1. Angiogenesis
The first description of the process of angiogenesis was made in 1794 by a British surgeon and anatomist, John Hunter [1]. Angiogenesis refers to the formation of new blood vessels from pre-existing ones [2] and requires a series of complex remodeling processes such as: vasodilation, cellular permeability, peri-endothelial support, proliferation, migration, lumenization, survival, differentiation and remodeling [3]. Triggering angiogenesis requires a disruption between the fine balance of pro-angiogenic and anti-angiogenic molecules [4]. The angiogenic process occurs during embryonic development [3], and, in the adult, it can occur in physiological situations, like the oestrus cycle in the females [5], or in wound healing situations [6], whereas in adult pathological scenarios it occurs in tumor growth [7], among others [8].
2. Notch Signaling Pathway in Angiogenesis
The angiogenic process is regulated by many signaling pathways. Among them is the Notch signaling pathway, an evolutionarily conserved signaling system that regulates proliferation, differentiation, cell-fate determination, progenitor and stem-cell self-renewal, in both embryonic and adult tissues [9][10]. The Notch signaling pathway was first discovered about 100 years ago when John Dexter, in 1904, described Drosophila melanogaster variants displaying wing phenotypes now associated with Notch pathway mutations [11]. Three years later, Thomas Morgan was able to identify the mutant alleles [12], but it was only after the molecular biology revolution, that Spyros Artavanis-Tsakonas and Michael Young were able to clone the Notch receptor and thus attribute the wing-notching phenotype to gene haplo-insufficiency [13][14]. These initial studies created the basic foundation for a new era in various fields, including developmental and stem cell biology, neuroscience, and cancer biology [15]. Since then, the Notch signaling pathway has been extensively characterized in its role in cell-fate determination, differentiation, proliferation, progenitor and stem-cell self-renewal, in a diversity of embryonic and adult tissues [10][16].
The Notch pathway is composed of five ligands (Jagged1, Jagged2, and Delta-like 1, 3, and 4) and four receptors (Notch 1–4). Notch protein receptors reside on the cell surface as non-covalently linked heterodimers that are comprised of the extracellular and transmembrane (intracellular) Notch polypeptides. Extracellular portions are characterized by numerous epidermal growth factor (EGF)-like repeats. Transmembrane portions include the membrane-proximal RBP-J-associated molecule (RAM) domain, which mediates interaction with several cytosolic and nuclear proteins; the Ankyrin (ANK) domain, which is also important for protein–protein interactions; two nuclear-localization sequences (NLSs); a carboxy-terminal transactivation domain (TAD), which is important for activating transcription; and a PEST (proline-, glutamate-, serine- and threonine-rich) domain, which is important for regulating Notch degradation. Transmembrane Notch3 and Notch4 are shorter and lack the TAD. The heterodimerization domain (HD) spans the region of interaction between the extracellular and transmembrane portions. The Notch ligands contain an EGF-like repeat region and a conserved sequence also known as Delta/Serrate/Lag (DSL). Jagged1 and Jagged2 each have a conserved cysteine-rich (CR) domain [10].
The functional Notch receptors are translocated to the cell surface as processed heterodimers. The final heterodimeric form of the receptors is preceded by a series of transformations which include: a Furin-dependent cleavage (S1 cleavage) in the Notch extracellular domain (NECD), that occurs during trafficking through the Golgi complex [17]; and a glycosylation by O-fucosyltransferase and Fringe family N-acetylglucosaminidyl transferases that is crucial for proper folding of the Notch receptor and the interaction with ligand-specific DSL domains (Delta, Serrate, Lag-2) [18].
Distinct ligand affinities exist for the various receptors, altered by glycosylation, which influences downstream transcriptional activation [19]. Classical Notch pathway activation requires ligand-receptor in adjacent cells because the ligands remain immobilized as transmembrane proteins. After Notch receptor binding, the ligand undergoes endocytosis into the ligand-presenting cell, which causes a mechanical disruption of the Notch receptor by changing the conformation of the negative regulatory region of the receptor. This conformational change in the Notch receptors allows for a second cleavage (S2) of the ectodomain by an Adam17 metalloprotease/Tnf-α converting enzyme (TACE) [20], followed by a third cleavage (S3) mediated by the presenilin-γ-secretase complex [21]. This sequence of cleavages leads to the release of the intracellular portion of the Notch receptor (NICD). The NICD contains nuclear localization signals (NLSs) within the RAM domain, which allows for the the translocation to the nucleus where it forms a complex with the inactive DNA-binding factor CSL/RBPjk (CBF1/Suppressor of Hairless/Lag1) and recruits other co-activator proteins from the Mastermind-like family of proteins such as MAML1 [22][23]. In the absence of NICD, RBP-Jk associates with a corepressor complex and acts as a transcriptional repressor of Notch target genes [24]. In turn, the NICD/RBP-Jk complex leads to the transcription of Notch downstream target genes, such as several helix–loop–helix transcription factors (Hey and Hes gene families among others) [10].
The study of Notch pathway components, specifically loss-of-function mouse mutants, has provided detailed information regarding the importance of these genes in the regulation of embryonic angiogenesis. Notch 1 is the most broadly studied Notch receptor and the main receptor responsible for Notch signaling associated phenotypes. Genetic deletion of Notch1 in mice results in embryonic lethality by severe vascular and cardiovascular defects [25]. The Notch2 gene was the second of the mammalian Notch family receptors to be cloned [26]. Later, mice homozygous for a hypomorphic Notch2 mutation were reported to present defects in development of the kidney, heart and eye vasculature [27]. Notch2 was also shown to be expressed in vascular smooth muscle cells and to play a critical role in vascular maturation [28][29][30]. Notch3 loss-of-function in mice resulted in profound structural and functional defects in arteries, due to impaired vascular maturation indicating a potential role in smooth muscle cell differentiation [31]. Notch4 is primarily expressed on the endothelium and the endocardium [32] and genetic deletion of Notch4 exacerbated the embryonic lethal vascular defects associated with Notch1, even though it did not produce a detectable phenotype on its own [33][34], suggestive of an important role in vascular development. Dll1 was shown to be essential for post-natal arteriogenesis [35] and established as a critical endothelial Notch ligand required for maintaining arterial identity during mouse fetal development [36]. Jagged1 is expressed in endothelial and vascular smooth muscle cells [37]. Jag1-null mouse mutants die at 11.5 dpc because of heart defects and abnormal development of the yolk sac and head vasculature [38]. Dll4 is the most broadly studied Notch ligand in vascular biology. The Dll4 ligand was first described as a vascular endothelium specific ligand [39]. In the developing embryo, expression of Dll4 is initially restricted to large arteries, whereas in adult mice its expression is limited to small arteries and capillaries [40]. Haplo-insufficiency of Dll4 in mice resulted in embryonic lethality at approximately 10.5 dpc due to defective vascular development, including abnormal stenosis and atresia of the aorta, defective arterial branching from the aorta, arterial regression, gross enlargement of the pericardial sac and failure to remodel the yolk sac vasculature. These studies revealed Dll4 to be essential for arterial patterning and vascular remodeling during embryonic development [40][41][42].
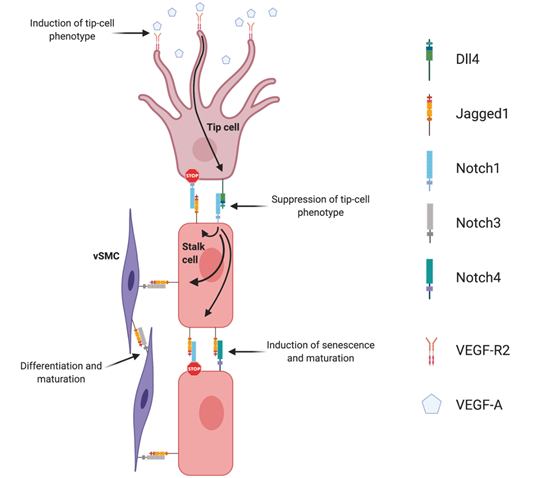
Sprouting angiogenesis (Figure 1) is strictly regulated by the interplay between VEGF and Dll4/Notch signaling. This interplay is the basis for the lateral induction model, currently accepted as the prevailing mechanistic model explaining sprouting angiogenesis, and tip- and stalk cell selection. The supporting evidence for this was established in the post-natal retina developing vascular plexus. In response to spatial gradients of Vegfa, secreted by neuroglia cells migrating radially ahead of the vascular front, tip-cells sprout filopodia towards this gradient. This effect is mediated by the interaction of Vegfa with Vegfr2 receptor, the concentration of which is especially high in tip-cells. Once tip-cells are selected and begin to move forward, formation of new capillaries begins because of the proliferation and migration of adjacent stalk ECs. When Vegfa gradients activate endothelial cells, they induce expression of Dll4 and Notch1 [43]. The tip-cell specific characteristics are preferably acquired by endothelial cells devoid of Notch1 and with high Dll4 expression. Dll4/Notch-associated transduction causes inhibition of sprouting by lowering ECs sensitivity to Vegfa. It was shown that in Dll4-hyperexpressing endothelial cells, expression of Vegfr2 was significantly inhibited [44]. Therefore, endothelial cells expressing Notch1 receptor, which was activated by adjacent Dll4 ligand, are prevented from transitioning to an active state, by lowering Vegfr2 levels, and thus Dll4/Notch signaling restricts the emergence of an excessive number of tip-cells, restricting excessive sprouting [45][46][47]. Specification of the tip/stalk cell phenotype by Notch is complex. In fact, even though Dll4 is the only ligand expressed in tip cells, Jagged1 and Dll1 are present in stalk cells [48]. Soluble Jagged1 was shown to reduce tip cell number, filopodia, and vessel density [47][49]. Moreover, Jagged1 was shown to have an opposite effect of Dll4 on branching morphogenesis, promoting endothelial cell proliferation and sprouting [50]. By using conditional Jag1 loss- and gain-of-function mouse mutants, it was demonstrated that in contrast to Dll4, Jagged1 is proangiogenic and functions by downregulating Dll4–Notch signaling. In this study, it was shown that Jagged1 function is of particular importance in stalk cells, where Jagged1 levels are high and therefore efficiently antagonize the potent Dll4 ligand. Consequently, it confers upon stalk cells little ability to activate Notch in adjacent tip cells. Jagged1 was also demonstrated to counteract Dll4–Notch signaling interactions between stalk ECs, which helped to sustain elevated VEGF receptor expression at the angiogenic front. Therefore, in this region, ECs were still able to respond to VEGF, which, in turn, promoted proliferation and the emergence of new tip cells. These authors have additionally proposed that Fringe-mediated modification of Notch is of critical importance in regulating tip cell selection. These results were later confirmed in wound healing angiogenesis, also revealing that Jagged1 is expressed downstream of Dll4–Notch1 in stalk cells, creating a negative feedback loop that blocks tip-cell Notch1 from being activated [51]. The authors also found that stalk EC Jagged1 could activate both Notch4 in other stalk ECs as well as Notch3 in neighboring smooth muscle cells, positively driving differentiation of vSMCs and maturation of the nascent vascular network [51].
Figure 1. Notch signaling regulation of angiogenesis. Endothelial Dll4, mainly expressed in tip cells, activates Notch1 in adjacent stalk cells leading to the up-regulation of endothelial Jagged1. Jagged1 antagonizes Dll4 ability to bind to activate Notch1 in tip cells, creating a negative feedback loop in the regulation of endothelial branching. Endothelial Jagged1 positively regulates vascular maturation by two possible mechanisms: by activating endothelial Notch4 in stalk cells and Notch3 in vascular smooth muscle cells (vSMC).
References
- Hunter, J. A Treatise on the Blood, Inflammation, and Gun-Shot Wounds. Clin. Orthop. Relat. Res. 1794.
- Flamme, I.; Frölich, T.; Risau, W. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J. Cell Physiol. 1997, 173, 206–210.
- Carmeliet, P. Angiogenesis in life, disease and medicine. Nature 2005, 438, 932–936.
- Karamysheva, A.F. Mechanisms of angiogenesis. Biochemistry 2008, 73, 751.
- Fraser, H.M.; Lunn, S.F. Angiogenesis and its control in the female reproductive system. Br. Med. Bull. 2000, 56, 787–797.
- Arbiser, J.L. Angiogenesis and the skin: A primer. J. Am. Acad. Dermatol. 1996, 34, 486–497.
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186.
- Carmeliet, P. Angiogenesis in health and disease. Nat. Med. 2003, 9, 653–660.
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776.
- Schweisguth, F. Regulation of Notch Signaling Activity. Curr. Biol. 2004, 14, R129–R138.
- Dexter, J.S. The analysis of a case of continuous variation in drosophila by a study of its linkage relations. Am. Nat. 1914, 48, 712.
- Morgan, T.H. The Theory of the Gene. Am. Nat. 1917, 51, 513.
- Wharton, K.A.; Johansen, K.M.; Xu, T.; Artavanis-Tsakonas, S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 1985, 43, 567–581.
- Kidd, S.; Kelley, M.R.; Young, M.W. Sequence of the notch locus of Drosophila melanogaster: Relationship of the encoded protein to mammalian clotting and growth factors. Mol. Cell. Biol. 1986, 6, 3094–3108.
- Fortini, M.E. Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 2009, 16, 633–647.
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012, 13, 654–666.
- Logeat, F.; Bessia, C.; Brou, C.; LeBail, O.; Jarriault, S.; Seidah, N.G.; Israël, A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 1998, 95, 8108–8112.
- Rana, N.A.; Haltiwanger, R.S. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 2011, 21, 583–589.
- Ntziachristos, P.; Lim, J.S.; Sage, J.; Aifantis, I. From fly wings to targeted cancer therapies: A centennial for notch signaling. Cancer Cell 2014, 25, 318–334.
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.R.; Cumano, A.; Roux, P.; Black, R.A.; Israël, A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216.
- De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S.; Schroeter, E.H.; Schrijvers, V.; Wolfe, M.S.; Ray, W.J.; et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 1999, 398, 518–522.
- Nam, Y.; Sliz, P.; Pear, W.S.; Aster, J.C.; Blacklow, S.C. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 2103–2108.
- Wilson, J.J.; Kovall, R.A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 2006, 124, 985–996.
- Kao, H.Y.; Ordentlich, P.; Koyano-Nakagawa, N.; Tang, Z.; Downes, M.; Kintner, C.R.; Evans, R.M.; Kadesch, T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998, 12, 2269–2277.
- Swiatek, P.J.; Lindsell, C.E.; Amo, F.F.D.; Weinmaster, G.; Gridley, T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994, 8, 707–719.
- Weinmaster, G.; Roberts, V.J.; Lemke, G. Notch2: A second mammalian Notch gene. Cell Death Differ. 1992, 116, 931–941.
- McCright, B.; Gao, X.; Shen, L.; Lozier, J.; Lan, Y.; Maguire, M.; Herzlinger, D.; Weinmaster, G.; Jiang, R.; Gridley, T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 2001, 128, 491–502.
- Varadkar, P.; Kraman, M.; Despres, D.; Ma, G.; Lozier, J.; McCright, B. Notch2 is required for the proliferation of cardiac neural crest-derived smooth muscle cells. Dev. Dyn. 2008, 237, 1144–1152.
- Hamada, Y.; Kadokawa, Y.; Okabe, M.; Ikawa, M.; Coleman, J.R.; Tsujimoto, Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 1999, 126, 3415–3424.
- Wang, Q.; Zhao, N.; Kennard, S.; Lilly, B. Notch2 and Notch3 Function Together to Regulate Vascular Smooth Muscle Development. PLoS ONE 2012, 7, e37365.
- Domenga, V.; Fardoux, P.; Lacombe, P.; Monet, M.; Maciazek, J.; Krebs, L.T.; Klonjkowski, B.; Berrou, E.; Mericskay, M.; Li, Z.; et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004, 18, 2730–2735.
- Uyttendaele, H.; Marazzi, G.; Wu, G.; Yan, Q.; Sassoon, D.; Kitajewski, J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 1996, 122, 2251–2259.
- Krebs, L.T.; Xue, Y.; Norton, C.R.; Shutter, J.R.; Maguire, M.; Sundberg, J.P.; Gallahan, D.; Closson, V.; Kitajewski, J.; Callahan, R.; et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000, 14, 1343–1352.
- Gridley, T. Notch signaling during vascular development. Proc. Natl. Acad. Sci. USA 2001, 98, 5377–5378.
- Limbourg, A.; Ploom, M.; Elligsen, D.; Sörensen, I.; Ziegelhoeffer, T.; Gossler, A.; Drexler, H.; Limbourg, F.P. Notch ligand Delta-like 1 is essential for postnatal arteriogenesis. Circ. Res. 2007, 100, 363–371.
- Sörensen, I.; Adams, R.H.; Gossler, A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 2009, 113, 5680–5688.
- Doi, H.; Iso, T.; Sato, H.; Yamazaki, M.; Matsui, H.; Tanaka, T.; Manabe, I.; Arai, M.; Nagai, R.; Kurabayashi, M. Jagged1-selective Notch Signaling Induces Smooth Muscle Differentiation via a RBP-Jκ-dependent Pathway. J. Biol. Chem. 2006, 281, 28555–28564.
- Xue, Y.; Gao, X.; Lindsell, C.E.; Norton, C.R.; Chang, B.; Hicks, C.; Gendron-Maguire, M.; Rand, E.B.; Weinmaster, G.; Gridley, T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 1999, 8, 723–730.
- Shutter, J.R.; Scully, S.; Fan, W.; Richards, W.G.; Kitajewski, J.; Deblandre, G.A.; Kintner, C.R.; Stark, K.L. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000, 14, 1313–1318.
- Duarte, A.; Hirashima, M.; Benedito, R.; Trindade, A.; Diniz, P.; Bekman, E.; Costa, L.; Henrique, D.; Rossant, J. Dosage-sensitive requirement for mouse Dll4 in artery development. Gene Dev. 2004, 18, 2474–2478.
- Krebs, L.T.; Shutter, J.R.; Tanigaki, K.; Honjo, T.; Stark, K.L.; Gridley, T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Gene Dev. 2004, 18, 2469–2473.
- Gale, N.W.; Dominguez, M.G.; Noguera, I.; Pan, L.; Hughes, V.; Valenzuela, D.M.; Murphy, A.J.; Adams, N.C.; Lin, H.C.; Holash, J.; et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA 2004, 101, 15949–15954.
- Liu, Z.J.; Shirakawa, T.; Li, Y.; Soma, A.; Oka, M.; Dotto, G.P.; Fairman, R.M.; Velazquez, O.C.; Herlyn, M. Regulation of Notch1 and Dll4 by vascular endothelial growth Factor in arterial endothelial Cells: Implications for modulating arteriogenesis and angiogenesis. Mol. Cell Biol. 2003, 23, 14–25.
- Williams, C.K.; Li, J.-L.; Murga, M.; Harris, A.L.; Tosato, G. Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood 2006, 107, 931–939.
- Siekmann, A.F.; Covassin, L.; Lawson, N.D. Modulation of VEGF signalling output by the Notch pathway. Bioessays 2008, 30, 303–313.
- Suchting, S.; Freitas, C.; le Noble, F.; Benedito, R.; Bréant, C.; Duarte, A.; Eichmann, A. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl. Acad. Sci. USA 2007, 104, 3225–3230.
- Hellström, M.; Phng, L.K.; Hofmann, J.J.; Wallgard, E.; Coultas, L.; Lindblom, P.; Alva, J.; Nilsson, A.K.; Karlsson, L.; Gaiano, N.; et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 2007, 445, 776–780.
- Roca, C.; Adams, R.H. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007, 21, 2511–2524.
- Sainson, R.C.A.; Aoto, J.; Nakatsu, M.N.; Holderfield, M.; Conn, E.; Koller, E.; Hughes, C.C. Cell-autonomous notch signaling regulates endothelial cell branching and proliferation during vascular tubulogenesis. FASEB J. 2005, 19, 1027–1029.
- Benedito, R.; Roca, C.; Sörensen, I.; Adams, S.; Gossler, A.; Fruttiger, M.; Adams, R.H. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 2009, 137, 1124–1135.
- Pedrosa, A.-R.; Trindade, A.; Fernandes, A.C.; Carvalho, C.; Gigante, J.; Tavares, A.T.; Diéguez-Hurtado, R.; Yagita, H.; Adams, R.H.; Duarte, A. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1134–1146.