
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Veronica Dodero | + 1570 word(s) | 1570 | 2020-11-05 08:31:40 | | | |
| 2 | Vicky Zhou | Meta information modification | 1570 | 2020-11-10 10:30:33 | | | | |
| 3 | Vicky Zhou | + 34 word(s) | 1604 | 2020-11-11 03:10:53 | | |
Video Upload Options
The self-assembly of proteins is an essential process for a variety of cellular functions including cell respiration, mobility and division. On the other hand, protein or peptide misfolding and aggregation is related to the development of Parkinson’s disease and Alzheimer’s disease, among other aggregopathies.
1. Introduction
Protein self-assembly plays a pivotal role in cellular physiology. It is a necessary process for the formation of any ordered protein structure [1]. Amelogenin proteins, which direct dental mineralization [2], are a remarkable example of self-assembled proteins that are found in structural tissues. On the other hand, self-organization and aggregation of proteins also takes part in physiopathological processes, as it is observed in different neurodegenerative diseases, where amyloid fibrils are deposited on different parts of the central nervous system [3]. The formation of protein aggregates has also become a fascinating research area, not only for mitigating diseases such as Alzheimer’s and Parkinson’s, but also in the pharmaceutical and food industries. In the pharmaceutical context, the uses of human recombinant proteins to treat a wide variety of diseases such as cancer, Hemophilia A and multiple sclerosis established the necessity to control and study the aggregation tendency of these formulations to avoid efficiency reduction and exacerbate immune response [4][5]. In this respect, a meticulous analysis of the therapeutic protein is needed in terms of protein structure, stability and oligomeric state. Moreover, in the food industry, the aggregation processes of whey proteins from milk [6], wheat gliadin and glutenin [7], and peanut proteins [8] are some examples that are continuously studied. The formed aggregates affect not only the organoleptic properties of the final product, but also its digestibility, bioavailability of amino acids and, in some cases, they can even induce or reduce inadequate immune responses to a toxic protein [9][10].
On the other hand, the aggregation capability of proteins from natural sources is used to generate nanoparticles for drug-delivery as in the case of whey, zeins and gliadins [11][12][13]. In this context, the use of techniques that allow the analysis of protein solutions to determine the presence of aggregates in a fast, reproducible and qualitative way is of key importance.
Nowadays, there is a wide range of possible methods for assessing protein and peptide self-assembly phenomena. However, to obtain molecular information, the use of specialized core facilities and also specific training on them is required. In this sense, spectroscopic methods like UV-Vis, fluorescence and circular dichroism together make a simple approach for evaluating proteins and peptides and assessing their self-assembly processes. In this regard, these spectroscopic techniques provide unique information in the case of complex systems, such as those composed by a mixture of proteins, where their analysis by sophisticated techniques as NMR (Nuclear Magnetic Resonance) is not easy to implement. The simplicity of these techniques in terms of implementation and analysis and their availability in multidisciplinary laboratories make them the first choice in the evaluation of protein and peptide self-assembly. Because of these reasons, we chose them as the topic of this review. It is worthy of mentioning that NMR is one of the most powerful tools for providing information about protein structure, dynamic and self-association at the atomic level, and is outside the scope of this review. The system needs to be labelled with 15N and 13C, and the broadening of the spectra peaks occurs when the size of the protein is above 30 kDa. As a consequence, methodologies such as Transverse relaxation-optimized spectroscopy (TROSY), methyl-TROSY and Cross-correlated relaxation-enhanced polarization transfer (CRINEPT) [14][15] have been developed to analyze self-assembled systems with a molecular mass above 30 kDa. Another NMR approach for studying supramolecular systems is solid-state nuclear magnetic resonance (ssNMR), which provides structural and dynamical information of complex biological systems especially in the case of protein aggregates and fibrils, such as the ones observed in amyloids [16][17][18]. For a deeper understanding of this technique, we suggest assessing the works cited here and some more specialized bibliography [19][20].
2. Protein Structure and Self-Assembly
Proteins and peptides develop a variety of activities in the cell. These molecules adopt a local folding referred to as the secondary structure (i.e., α-helix or β-sheet structure) and a tridimensional location of the secondary structure in space, known as the tertiary structure. Moreover, some proteins adopt a quaternary structure, which implicates the association between protein subunits and their arrangements from dimers to oligomers. [21][22]. The self-association of proteins in different oligomeric states is mainly governed by nonbonded interactions such as the Van der Waals forces, hydrogen and ionic bonds and π–π interactions [23]. However, in some cases, the formation of specific disulfide bonds is critical for protein oligomerization [21]. The size of the self-associated superstructures could range from nm to µm. Some systems are stabilized in the solution, forming a dispersion; meanwhile, others form insoluble amorphous aggregates or ordered ones, like fibrils [24].
Amorphous aggregates as spherules and fractal-like clusters can be detected in the first stages of the self-assembly process, as in the case of the 33-mer gliadin peptide related to celiac disease [25] or the silk protein sericin [26]. In the case of fibrils, they are classified as non-amyloid and amyloids. Representative non-amyloid fibrillar structures are mainly related to motility and scaffold functions in the cell, such as actin fibrils, microtubules, collagen, among others [27]. On the other hand, amyloid fibrils are well known to be a hallmark of neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and type II diabetes, among other pathologies [3].
Recently, it was demonstrated that colloidal protein behavior has an essential role in self-assembly processes, as occurs in condensation and Ostwald ripening [28][29]. Ostwald ripening explains the formation of protein droplets in a liquid system, such as the nucleolus or the cytoplasm, and is of great importance in cellular physiology and stress response [30]. Interestingly, there are self-associated proteins that form regular spatial patterns. One of the most well-known is the formation of superstructures like viral capsids, which require a specific number of monomers to generate the self-associated system. In particular, conditions of salts, temperature and pH, proteins could also associate to form crystals. This phenomenon is of particular interest because it allows the 3D structure resolution of the protein by X-ray diffraction [31]. All the structures presented are shown in Figure 1.
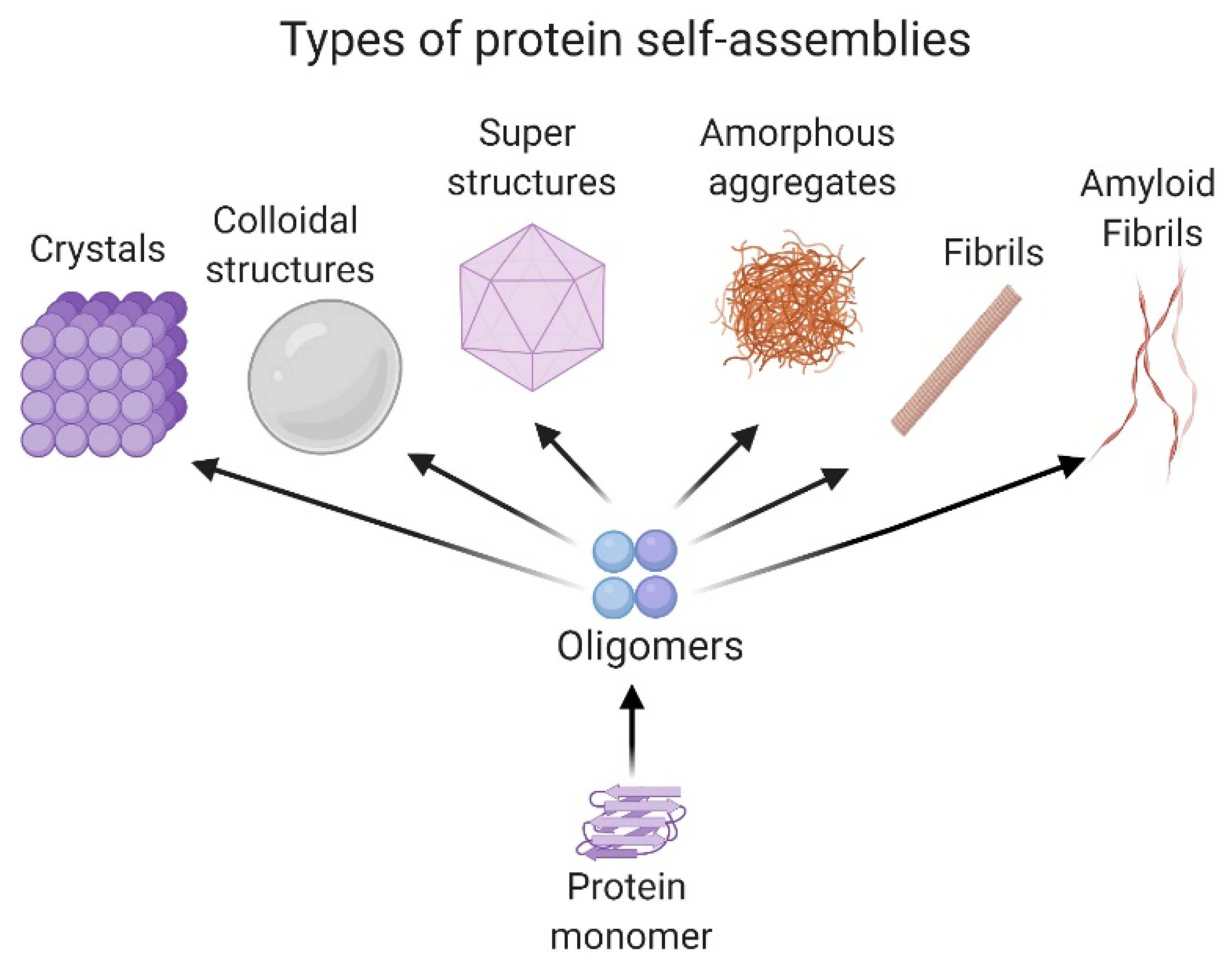
Secondary structure conversion of peptides and proteins from native conformation towards a β-sheet structure, independently of the protein sequence, has been described in a variety of proteins and especially in the amyloidogenic ones. In this case, non-branched fibrils are formed with a β-sheet conformation named cross-β, where the hydrogen bonding direction is parallel to the fiber axis, and the β-strands are perpendicular, like the rungs of a ladder [32]. Recently, it was pointed out that the α-sheet structure, referred to as “polar pleated sheet”, has an essential role in the formation of toxic oligomers. This structure is highly similar to the β-sheet, except that the carbonyl oxygen atoms are aligned on one face of a strand and the NH groups on the other, instead of alternating with each other, giving rise to different physical properties to the protein. This structure was found in the early stages of the Aβ-peptide [33][34] and the transthyretin protein (TTR) aggregation [35].
Moreover, another relevant motif for protein self-assembly is Polyproline II (PPII). This secondary structure is a left-handed helix that does not depend on the formation of hydrogen bonds in the backbone or salt bridges, but regularly establishes hydrogen bonds with the solvent [36]. This structure has been shown to be highly abundant, especially in structural proteins like collagen and exposed protein segments [37][38]. Additionally, it can interconvert into other forms such as β-turns and β-strands because of the proximity of the corresponding dihedral angles [39]. PPII plays a vital role in protein–protein interactions [40] and aggregation, as it was detected as the intermediate motif during the self-aggregation of lysozyme [41], the Aβ-peptide [42] and gliadin peptides [43][44].
3. Some Considerations in Order to Avoid Protein Aggregation
Apart from aggregation and self-assembly studies, it might be of interest to study the monomeric form of the different proteins with prone aggregation or self-assembly tendencies. To perform this kind of study, different strategies could be applied, depending on the protein under investigation. The agents that may be used for these purposes are extensive, with chaotropes, aminoacids, detergents, sugars and polyhydric alcohols, and polymers being among the most used. The mechanisms by which these agents avoid aggregation are diverse, and in some cases are not entirely understood. Some of these agents may decrease the rate of protein association and dissociation by being kept out from the protein–protein encounter surface, with the condition of not interacting with the protein monomer [45]. For instance, in the works on the ISD11 protein (part of the Fe-S cluster mitochondrial supercomplex in eukaryotes), SDS (0.45 mM) and DDM (1 mM) were used to help the protein refolding and prevent aggregation [46]. Another interesting example to avoid aggregation is the addition of glucose (up to 5%) in the protein buffer (apart from the buffer agent, salts, and any other co-solvents) of the UDK-c protein from D. melanogaster [47]. In a more biotechnological approach, there are many examples of E. coli acting as a chaperone of co-expression to obtain soluble proteins [48]. It is important to mention that, in general, the combination of co-solvents is useful to avoid aggregation [49]. In addition to the decision as to the type and amount of co-solvent to be used, it is relevant to take into consideration the kind of experiment that we are attempting to perform afterwards, i.e., we should avoid using high salts and glycerol concentrations to perform functional studies including DNA and DNA binding proteins [47].
References
- Chiesa, G.; Kiriakov, S.; Khalil, A.S. Protein assembly systems in natural and synthetic biology. BMC Biol. 2020, 18, 35.
- Engelberth, S.A.; Bacino, M.S.; Sandhu, S.; Li, W.; Bonde, J.; Habelitz, S. Progression of Self-Assembly of Amelogenin Protein Supramolecular Structures in Simulated Enamel Fluid. Biomacromolecules 2018, 19, 3917–3924.
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773.
- Ratanji, K.D.; Derrick, J.P.; Dearman, R.J.; Kimber, I. Immunogenicity of therapeutic proteins: Influence of aggregation. J. Immunotoxicol. 2014, 11, 99–109.
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat Rev. Drug Discov. 2008, 7, 21–39.
- Wolz, M.; Mersch, E.; Kulozik, U. Thermal aggregation of whey proteins under shear stress. Food Hydrocoll. 2016, 56, 396–404.
- Ma, S.; Han, W.; Li, L.; Zheng, X.; Wang, X. The thermal stability, structural changeability, and aggregability of glutenin and gliadin proteins induced by wheat bran dietary fiber. Food Funct. 2019, 10, 172–179.
- Sun, X.; Jin, H.; Li, Y.; Feng, H.; Liu, C.; Xu, J. The Molecular Properties of Peanut Protein: Impact of Temperature, Relative Humidity and Vacuum Packaging during Storage. Molecules 2018, 23, 2618.
- Singh, T.K.; Oiseth, S.K.; Lundin, L.; Day, L. Influence of heat and shear induced protein aggregation on the in vitro digestion rate of whey proteins. Food Funct. 2014, 5, 2686–2698.
- Satitsuksanoa, P.; Jansen, K.; Globinska, A.; van de Veen, W.; Akdis, M. Regulatory Immune Mechanisms in Tolerance to Food Allergy. Front. Immunol. 2018, 9, 2939.
- Islam, M.S.; Reineke, J.; Kaushik, R.; Woyengo, T.; Baride, A.; Alqahtani, M.S.; Perumal, O. Bioadhesive Food Protein Nanoparticles as Pediatric Oral Drug Delivery System. ACS Appl. Mater. Interfaces 2019, 11, 18062–18073.
- Gulfam, M.; Kim, J.E.; Lee, J.M.; Ku, B.; Chung, B.H.; Chung, B.G. Anticancer drug-loaded gliadin nanoparticles induce apoptosis in breast cancer cells. Langmuir 2012, 28, 8216–8223.
- Ha, H.K.; Rankin, S.A.; Lee, M.R.; Lee, W.J. Development and Characterization of Whey Protein-Based Nano-Delivery Systems: A Review. Molecules 2019, 24, 3254.
- Sprangers, R.; Velyvis, A.; Kay, L.E. Solution NMR of supramolecular complexes: Providing new insights into function. Nat. Methods 2007, 4, 697–703.
- Riek, R.; Pervushin, K.; Wüthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000, 25, 462–468.
- Naito, A.; Kawamura, I. Solid-state NMR as a method to reveal structure and membrane-interaction of amyloidogenic proteins and peptides. Biochim. Biophys. Acta 2007, 1768, 1900–1912.
- Weingarth, M.; Baldus, M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc. Chem. Res. 2013, 46, 2037–2046.
- Linser, R. Solid-state NMR spectroscopic trends for supramolecular assemblies and protein aggregates. Solid State Nucl. Magn. Reson. 2017, 87, 45–53.
- Duer, M.J. Solid state NMR Spectroscopy: Principles and Applications; Blackwell Science Ltd.; John Wiley & Sons: Oxford, UK, 2008.
- Cavanagh, J.; Fairbrother, W.J.; Palmer III, A.G.; Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice; Academic Press; Elsevier: Burllington, NJ, USA, 2007.
- Liu, S. A review on protein oligomerization process. Int. J. Precis. Eng. Manuf. 2015, 16, 2731–2760.
- Koehl, P. Protein Structure Classification. In Reviews in Computational Chemistry; Lipkowitz, K.B., Cundari, T.R., Gillet, V.J., Boyd, D.B., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2006; Chapter 1; pp. 1–55.
- Dhotel, A.; Chen, Z.; Delbreilh, L.; Youssef, B.; Saiter, J.M.; Tan, L. Molecular motions in functional self-assembled nanostructures. Int. J. Mol. Sci. 2013, 14, 2303–2333.
- van der Linden, E.; Venema, P. Self-assembly and aggregation of proteins. Curr. Opin. Coll. Interface Sci. 2007, 12, 158–165.
- Herrera, M.G.; Benedini, L.A.; Lonez, C.; Schilardi, P.L.; Hellweg, T.; Ruysschaert, J.M.; Dodero, V.I. Self-assembly of 33-mer gliadin peptide oligomers. Soft Matter 2015, 11, 8648–8660.
- Khire, T.S.; Kundu, J.; Kundu, S.C.; Yadavalli, V.K. The fractal self-assembly of the silk protein sericin. Soft Matter 2010, 6, 2066–2071.
- van Raaij, M.J. and Mitraki, A. Natural Fibrous Proteins: Structural Analysis, Assembly, and Applications. In Proteins in Solution and at Interfaces; Ruso, J.M., Piñeiro, Á., Eds.; Jon Wiley & Sons: Hoboken, NJ, USA, 2013; Chapter 11; pp. 219–232.
- Crick, S. and Pappu, R. Thermodynamic and Kinetic Models for Aggregation of Intrinsically Disordered Proteins. In Peptide Folding, Misfolding, and Nonfolding; Schweitzer-Stenner, R., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 413–440.
- Dine, E.; Gil, A.A.; Uribe, G.; Brangwynne, C.P.; Toettcher, J.E. Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Syst. 2018, 6, 655–663.
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435.
- McManus, J.J.; Charbonneau, P.; Zaccarelli, E.; Asherie, N. The physics of protein self-assembly. Current Opin. Coll. Interface Sci. 2016, 22, 73–79.
- Rambaran, R.N.; Serpell, L.C. Amyloid fibrils: Abnormal protein assembly. Prion 2008, 2, 112–117.
- Shea, D.; Hsu, C.C.; Bi, T.M.; Paranjapye, N.; Childers, M.C.; Cochran, J.; Tomberlin, C.P.; Wang, L.; Paris, D.; Zonderman, J.; et al. alpha-Sheet secondary structure in amyloid beta-peptide drives aggregation and toxicity in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2019, 116, 8895–8900.
- Meng, F.; Lu, T.; Li, F. Stabilization of Solvent to alpha-Sheet Structure and Conversion between alpha-Sheet and beta-Sheet in the Fibrillation Process of Amyloid Peptide. J. Phys. Chem. B 2019, 123, 9576–9583.
- Childers, M.C.; Daggett, V. Drivers of alpha-Sheet Formation in Transthyretin under Amyloidogenic Conditions. Biochemistry 2019, 58, 4408–4423.
- Whittington, S.J.; Creamer, T.P. Salt bridges do not stabilize polyproline II helices. Biochemistry 2003, 42, 14690–14695.
- Berisio, R.; Loguercio, S.; De Simone, A.; Zagari, A.; Vitagliano, L. Polyproline helices in protein structures: A statistical survey. Protein Pept. Lett. 2006, 13, 847–854.
- Adzhubei, A.A.; Sternberg, M.J. Left-handed polyproline II helices commonly occur in globular proteins. J. Mol. Biol. 1993, 229, 472–493.
- Bochicchio, B.; Tamburro, A.M. Polyproline II structure in proteins: Identification by chiroptical spectroscopies, stability, and functions. Chirality 2002, 14, 782–792.
- Cubellis, M.V.; Caillez, F.; Blundell, T.L.; Lovell, S.C. Properties of polyproline II, a secondary structure element implicated in protein-protein interactions. Proteins 2005, 58, 880–892.
- Blanch, E.W.; Morozova-Roche, L.A.; Cochran, D.A.; Doig, A.J.; Hecht, L.; Barron, L.D. Is polyproline II helix the killer conformation? A Raman optical activity study of the amyloidogenic prefibrillar intermediate of human lysozyme. J. Mol. Biol. 2000, 301, 553–563.
- Eker, F.; Griebenow, K.; Schweitzer-Stenner, R. Abeta (1-28) fragment of the amyloid peptide predominantly adopts a polyproline II conformation in an acidic solution. Biochemistry 2004, 43, 6893–6898.
- Herrera, M.G.; Zamarreno, F.; Costabel, M.; Ritacco, H.; Hutten, A.; Sewald, N.; Dodero, V.I. Circular dichroism and electron microscopy studies in vitro of 33-mer gliadin peptide revealed secondary structure transition and supramolecular organization. Biopolymers 2014, 101, 96–106.
- Herrera, M.G.; Gomez Castro, M.F.; Prieto, E.; Barrera, E.; Dodero, V.I.; Pantano, S.; Chirdo, F. Structural conformation and self-assembly process of p31-43 gliadin peptide in aqueous solution. Implications for celiac disease. FEBS J. 2020, 287, 2134–2149.
- Baynes, B.M.; Trout, B.L. Rational design of solution additives for the prevention of protein aggregation. Biophys. J. 2004, 87, 1631–1639.
- Herrera, M.G.; Pignataro, M.F.; Noguera, M.E.; Cruz, K.M.; Santos, J. Rescuing the Rescuer: On the Protein Complex between the Human Mitochondrial Acyl Carrier Protein and ISD11. ACS Chem. Biol. 2018, 13, 1455–1462.
- Bondos, S.E.; Bicknell, A. Detection and prevention of protein aggregation before, during, and after purification. Anal. Biochem. 2003, 316, 223–231.
- Kumat, T.; Samuel, D.; Jayaraman, G.; Srimathi, T.; Yu, C.J. The role of proline in the prevention of aggregation during protein folding in vitro. IUBMB Life 1998, 46, 509–517.
- Lu, H.; Zhang, H.; Wang, Q.; Yuan, H.; He, W.; Zhao, Z.; Li, Y. Purification, refolding of hybrid hIFNgamma-kringle 5 expressed in Escherichia coli. Curr. Microbiol. 2001, 42, 211–216.




