| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jana Jankovičová | + 5697 word(s) | 5697 | 2020-10-19 12:12:51 | | | |
| 2 | Jana Jankovičová | Meta information modification | 5697 | 2020-10-22 14:43:47 | | | | |
| 3 | Camila Xu | Meta information modification | 5697 | 2020-10-26 09:27:29 | | | | |
| 4 | Camila Xu | + 1 word(s) | 5698 | 2020-11-02 10:57:01 | | |
Video Upload Options
Tetraspanins are transmembrane proteins with ability to interact with each other and with other non-tetraspanin partners, building up a complex called tetraspanin web. This dynamic structure participates in many cellular processes. Although currently, the tetraspanin proteins found in extracellular vesicles are mostly applied as markers, increasing evidence points to their role in extracellular vesicle biogenesis, cargo selection, cell targeting, and cell uptake under both physiological and pathological conditions.
1. Introduction
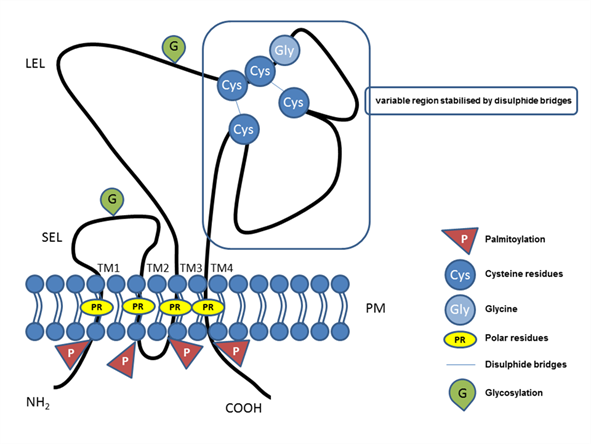
The tetraspanin protein family includes more than 30 members, and tetraspanins have been found on the plasma membrane or in endosomal or lysosomal compartments of almost all cell types [1]. The tetraspanin family includes distinct proteins characterized by their common specific molecular structure (Figure 1), represented by four transmembrane domains containing conserved polar residues; a small extracellular loop (SEL) and large extracellular loop (LEL) with 4, 6, 7 or 8 conserved cysteine residues; and short cytoplasmic tails [2][3][4][5][6]. Polar residues stabilize the tertiary structure [7]. Tetraspanins are usually post-translationally modified by glycosylation in extracellular domains and palmitoylation at intracellular cysteines [1]. They also contain a tyrosine-based sorting motif for intracellular compartment targeting that may mediate internalization via associated proteins [8]. Tetraspanins are generally accepted to have an important role as organizers of each other and of distinct transmembrane and cytosolic proteins (integrins, members of the immunoglobulin superfamily, proteases) into a multimolecular membrane network called the tetraspanin web [6][9][10][11][12]. In general, tetraspanins are implicated in many cellular processes, such as cell adhesion [13], regulation of cell motility, and/or morphology, fusion, signaling, and other functions [10][14][15][16][17].
Figure 1. Illustrative schema of tetraspanin structure. Tetraspanin proteins traverse the plasma membrane (PM) four times, thus, defining the four transmembrane domains (TM1, TM2, TM3, TM4) with conserved polar residues (PRs) in their structure. In the extracellular space, the small (SEL) and large (LEL) extracellular loops can be recognized. The LEL contains a highly conserved CCG motif and possibly an additional two, four, five, six, or eight conserved cysteine residues (Cys). Between the cysteine residues, two disulfide bridges that enable folding of the LEL can be formed. Tetraspanins are post-translationally modified by glycosylation (G) in the large or small extracellular domain and palmitoylation (P) at the intracellular cysteine residues. Short N-terminal and C-terminal tails are oriented intracellularly.
2. Extracellular vesicles
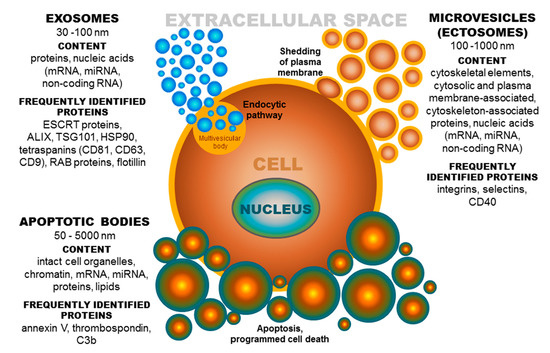
Based on published data, the term extracellular vesicles (EVs) denotes a population of different groups of vesicles classified according to their biogenesis and release pathway, evidently overlapping in some cases. Individual membrane vesicle categories differ, not only in origin, size, and morphology, but also in content [18] (Figure 2). The currently known data regarding the molecular cargo of extracellular vesicles (lipids, RNAs, and proteins) are summarized in the ExoCarta database [19][20][21] and in Vesiclepedia, a compendium for EVs [22][23]. Based on their diameter, EVs can be classified into several groups, namely, ectosomes, or shedding microvesicles (MVs) (100–1000 nm) [24][25][26]; exosomes (EXs) (30–100 nm) [25]; apoptotic bodies (ABs) (50–5000 nm) [27][28]; and other EV subsets, as reported by Shah et al. [29]. These groups of extracellular vesicles also differ in their origin [30]. Exosomes have an endocytic origin and are released from multivesicular endosomes. The biogenesis of EXs begins with the internalization of molecules via endocytosis [31]. Subsequently, endocytosed molecules are either recycled to the plasma membrane or trafficked to multivesicular bodies [18]. Multivesicular bodies fuse with the plasma membrane, leading to the release of intraluminal vesicles as exosomes into the extracellular microenvironment [32]. Whereas, MVs are formed by blebbing of the plasma membrane and subsequent fission of the membrane blebs [18][33], ABs are shed from the membrane of cells during the process of programmed cell death or apoptosis [18][28].
Figure 2. Extracellular vesicles: Their origin, size, and cargo. ESCRT-endosomal sorting complex required for transport; ALIX-protein regulating cellular mechanisms, including endocytic membrane trafficking and cell adhesion; TSG101-tumor susceptibility gene 101 protein; HSP-heat shock protein, CD-cluster of differentiation; RAB-proteins included in regulation of endocytosis and secretory processes, C3b-complement component.
While it is generally accepted that the main function of EVs is the mediation of intercellular communication [34][35][36], they are also involved in cell homeostasis, coagulation, and waste management [33]. Although cells typically release several major EV populations defined by biophysical properties and biological functions, the heterogeneity among them is obvious and likely underlies the specific role of EV subpopulations in individual cellular processes [37]. EVs are effective intercellular transporters of proteins, lipids, and nucleic acids. The protein cargo of extracellular vesicles includes proteins participating in cell adhesion (integrins, ICAM (intracellular adhesion molecule)), intracellular trafficking (GTPases, RAB (proteins included in regulation of endocytosis and secretory processes, annexins), and signal transduction (protein kinases, G proteins, β catenin) [38]. EVs are usually enriched in tetraspanin proteins (mainly CD9, CD63, and CD81) and other proteins, such as ALIX (protein regulating cellular mechanisms), TSG101 (tumor susceptibility gene 101 protein), MHC1 (major histocompatibility complex 1), and HSP90 (heat shock protein 90) [39][40]. Regarding lipids, EVs are characterized by the presence of phosphatidylserine, cholesterol, sphingomyelins, and ceramides [41][42], which participate not only in intercellular signaling, but also ensure structural stability [43]. EVs may also carry nucleic acids (genomic DNA, mitochondrial DNA, mRNA, miRNA, and long non-coding RNA) [44]. Overall, EVs can alter the physiological and pathological function of recipient and parent cells through the transfer of proteins, lipids, and RNA [45]. Exosome uptake (and likely uptake of the other types of EVs) can cause activation, differentiation, or dedifferentiation of target cells depending on the delivered cargo [46]. Notably, it was shown that exosomal mRNA transferred to recipient cells can be translated and that miRNA may regulate gene expression in recipient cells [44]. Beside the somatic (body) systems, many tissues and cells of the reproductive tract release EVs, which are believed to participate in various steps of the reproduction process (reviewed in Reference [47]). The participation of tetraspanin family proteins, the most prevalent proteins in EVs [48][49][50], which are routinely used only as markers (mostly for exosomes) in mammalian fertilization, has been demonstrated [51][52][53].
3. Role of Tetraspanins in EV Formation
Exosomes can be formed through ESCRT (endosomal sorting complex required for transport)-dependent or ESCRT-independent pathways (reviewed in References [40][54][55][56]). Studies by van Niel et al. [56] and Chairoungdua et al. [57] indicated that tetraspanins may be crucial players in ESCRT-independent pathways of exosome biogenesis and secretion. It is well known that tetraspanins can modulate membranes by affecting their curvature [58][59][60][61], which directly predetermines their participation in EV formation. Considering the reversed cone-like molecular shape of CD9, Umeda et al. [59] suggested that clustering of tetraspanin molecules could modulate membrane curvature to enable exosome budding and facilitate subsequent interaction of the C-terminal region of tetraspanins (cytoplasmic tail) with cytoskeletal actin through ezrin, radixin and moesin proteins [62], which may be involved in EV fission from the parent cell membrane.
4. Role of Tetraspanins in EV Cargo Selection, Targeting, and Uptake
Although the processes of targeting and uptake (internalization of EVs by recipient cells) are currently not fully understood, it is evident that molecules involved in tetraspanin-enriched microdomains of EVs and TEMs of recipient cells, especially tetraspanins, integrins, and other adhesion proteins, play a key role in the process of binding, fusion, and targeting of extracellular vesicles and in selective uptake of EVs by recipient cells [37]. As shown in exosomes, the internalization of membrane molecules into endosome compartments is related to the rearrangement of TEMs [63]. Based on previous findings, Rana et al. [49] hypothesized that regulation of the protein assembly of exosomes, and potentially, the recruitment of microRNA is ensured by tetraspanins. In 2012, Rana et al. [50] reported for the first time that selective exosome uptake by cells and tissues is critically dependent on the tetraspanin web composition. Therefore, it seems that TEMs are involved not only in molecular internalization and recycling, but that tetraspanin proteins (at least CD9, CD81, CD82, CD63, and tetraspanin-8) also regulate the sorting of proteins and possibly RNA to EVs (reviewed in Reference [64]). A quantitative proteomic analysis conducted by Perez-Hernandez et al. [65] showed that the TEM interaction network corresponds to 45% of the exosomal proteome. Consequently, the diminishment of the tetraspanin profile of extravesicular TEMs can result in a decrease in the concentration of some of their associated partners. In general, common components of TEMs within the cell membrane are tetraspanins, integrins, and other adhesion receptors and transmembrane receptor proteins [10][15][66][67]. Therefore, changes in the expression of specific tetraspanins may modulate selective targeting and uptake of EVs, thereby affecting the cellular response. Willms et al. [37] suggested the utilization of the tetraspanin (and integrin) profile as a distinguishing criterion for individual EV subpopulations. The involvement of tetraspanins in vesicular cargo selection was also reported in neuroblastoma cells, where EVs targeted the cells depending on the presence of CD63 tetraspanin or amyloid precursor protein [42]. The exosomal tetraspanins CD9 and CD81, together with integrin αvβ3, were shown to be involved in the targeting and uptake of exosomes by dendritic cells, and a complex of tetraspanin-8 with integrin subunit CD49d (α4) determines the selective targeting and uptake of tumor-derived exosomes by endothelial cells [68]. In a very recent study, Umeda et al. [59] revealed the cone-like molecular shape of CD9 (based on the crystal structure of CD9 and cryo-electron microscopy of human CD9) and proposed two potential roles of tetraspanins in exosome function. Tetraspanin clustering may directly affect membrane curvature and thereby enable exosome budding, or alternatively, tetraspanins can control vesicular cargo sorting through association with other partner proteins via EWI family proteins [62,69], thereby establishing a complex functional molecular network [59]. To deliver proteins or nucleic acids, EVs undergo direct fusion with the target cell membrane or the endosomal compartment membrane after endocytic uptake [70][71][72][73]. The uptake of EVs can occur through several endocytic pathways, including clathrin-dependent endocytosis and clathrin-independent pathways, such as caveolin-mediated uptake, macropinocytosis, phagocytosis, and lipid raft-mediated internalization. The same authors stated that the different uptake mechanism of individual EV populations likely depends on the proteins and glycoproteins on the surface of the vesicle and the target cell (reviewed in Reference [72]). Based on the fact that tetraspanins are involved in many cellular processes, including vesicular and cellular fusion [10][74], they are ‘predestined’ to participate in EV binding and uptake by cells (reviewed in References [75][76][77][78]). In vitro and in vivo experiments have suggested that exosomes diffuse throughout the whole body, with selective enrichment in different cells/organs depending on exosome-tetraspanin complexes and cell ligands [50]. These findings were supported by experiments on somatic cells, where a reduction in EV uptake by target/recipient cells was observed after treatment with antibodies against tetraspanin-8, CD9, CD81, and CD151 [79][80][81]. It should be noted that in some cases, a cellular response, such as activation/inhibition of signaling pathways, may be induced by EVs without their internalization, as has been illustrated in several studies [82][83][84][85][86][87]. It is generally believed that tetraspanins act as regulators of the adhesive activity of several adhesion molecules, including integrins [88][89][90][91][92][93][94], and thus, they may not only control adhesion, but may also affect signaling pathways. Therefore, specific tetraspanin and integrin profiles of EVs could ensure the specific role of individual EV subpopulations.
5. Role of Tetraspanins in Immunostimulatory and Immunosuppressive Properties of EV Subpopulations in Cancer
In recent years, great effort has been devoted to the elucidation of EV function in cancer biology (reviewed in References [95][96]). Numerous EVs secreted by cancer cells transport their molecular content to various target cells, including endothelial, epithelial, and immune cells, and serve as regulators of intercellular communication of cancer cells [95][97]. Increasing experimental data indicate a crucial role of EVs in cancer progression. On the other hand, several studies have suggested that non-tumoral cells may suppress cancer initiation and progression via EVs (reviewed in Reference [95]). Extracellular vesicles can participate in remodeling of the tumor microenvironment, which affects endothelial cells, epithelial cells, stromal cells, fibroblasts, and macrophages [98][99][100]. Findings regarding tetraspanins and integrins are also related to their role in organotropism, the non-random distribution of metastases in an organ-specific pattern [50][98][101]. Hoshino et al. [101] suggested that integrins in EVs mediate not only their adhesion to recipient cells, but also trigger signaling and inflammatory responses in recipient cells, thereby leading organs to become permissive for metastatic cell growth. As reported by Yue et al. [98], the communication between tumor-derived EVs and the matrix is also significantly affected by the interaction of tetraspanins (CD151 and tetraspanin-8) with integrins, and proteases. In addition, EVs can mediate hematopoietic and stromal cell activation, including (lymph) angiogenesis, and stimulate epithelial-mesenchymal transition in neighboring non-metastatic tumor cells [98].
6. Role of EV Tetraspanins in Antigen Presentation
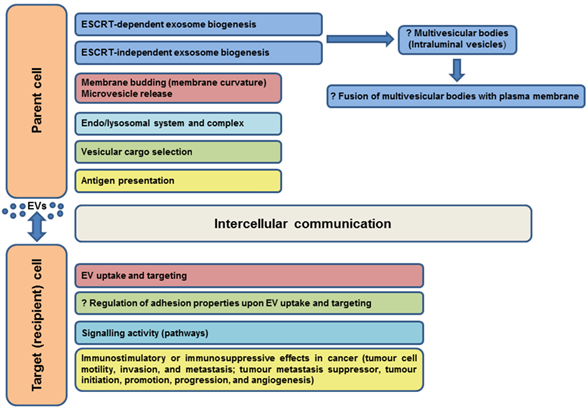
Most immune system cells (likely all), including antigen-presenting cells (APCs), secrete extracellular vesicles, and depending on the type and status of the parent cell, EVs may modulate the immune response in various ways (initiation, expansion, maintenance, or silencing) [102]. Professional antigen-presenting cells mainly include dendritic cells, macrophages, and B cells. It was shown that EVs released by APCs can induce an adaptive immune response by the distribution of MHC-peptide complexes and presentation of these antigens to specific T cells ( [34][103][104], as reviewed in Reference [64]). As known molecular organizers, tetraspanins present on the surface of immune cells have been shown to play an important role in the adaptive immune response via involvement in antigen presentation and formation of molecular complexes in immunological synapse assembly [74][105][106][107][108][109][110]. The interaction of the APC tetraspanins CD9, CD37, CD53, CD81, and CD82 with MHC molecules, as well as engagement of MHC-peptide complexes in TEMs, has led to the consideration of their role in the formation of MHC-II multimers and enhanced antigen presentation [107][111][112][113][114]. According to Kropshofer et al. [107], the organization of MHC-II complexes in TEMs could determine the composition of the immunological synapse, and subsequently, the quality of the T helper cell response. Andreu and Yáñez-Mó [64] suggested that in addition to regulation of the expression and sorting of MHC to EVs, tetraspanins may be responsible for the degree of MHC complex clustering in EV membranes sufficient to induce an immune response. Tetraspanins can also participate in the regulation of different steps in the immune response through their ability to alter the function of their associated partners (reviewed in Reference [102]). The evidenced or predicted involvements of tetraspanins in EV functions in somatic cells are summarized in Figure 3.
Figure 3. Evidenced/predicted involvement of tetraspanins in EV functions in somatic cells. EVs-extracellular vesicles, ESCRT-endosomal sorting complex required for transport; ?-predicted role.
7. Conclusion
Extensive research in the field of extracellular vesicles has shown their participation via their molecular cargo in many cellular functions in mammals. Although currently, tetraspanin proteins are applied mostly as markers of extracellular vesicles, increasing evidence points to their pivotal role in EV biogenesis, cargo selection, cell targeting, and uptake, which underlies their effect on distinct cellular processes under both physiological and pathological conditions. Understanding the enigmatic role of EV tetraspanins is challenging, due to their complexity. Their individual abundance differs among species, individuals, organs, and anatomical regions. Researchers working in the field of EV tetraspanins face many difficulties related to their function in multimolecular complexes. Their composition likely differs depending on the processes in which they are involved. In sum, the constantly updated knowledge regarding extracellular vesicles has provided substantial findings that markedly change the point of view on physiological, as well as pathological processes, and the contribution of EV tetraspanins should not be omitted.
References
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. Functional domains in tetraspanin proteins. Trends in Biochemical Sciences 2003, 28, 106–112, doi:10.1016/S0968-0004(02)00014-2.
- Berditchevski, F.; Odintsova, E.; Sawada, S.; Gilbert, E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002, 277, 36991–37000, doi:10.1074/jbc.M205265200.
- Charrin, S.; Manié, S.; Oualid, M.; Billard, M.; Boucheix, C.; Rubinstein, E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002, 516, 139–144, doi:10.1016/s0014-5793(02)02522-x.
- Yang, X.; Claas, C.; Kraeft, S.-K.; Chen, L.B.; Wang, Z.; Kreidberg, J.A.; Hemler, M.E. Palmitoylation of Tetraspanin Proteins: Modulation of CD151 Lateral Interactions, Subcellular Distribution, and Integrin-dependent Cell Morphology. Mol Biol Cell 2002, 13, 767–781, doi:10.1091/mbc.01-05-0275.
- Huang, S.; Yuan, S.; Dong, M.; Su, J.; Yu, C.; Shen, Y.; Xie, X.; Yu, Y.; Yu, X.; Chen, S.; et al. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics 2005, 86, 674–684, doi:10.1016/j.ygeno.2005.08.004.
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205, doi:10.1007/PL00000933.
- Tang, M.; Yin, G.; Wang, F.; Liu, H.; Zhou, S.; Ni, J.; Chen, C.; Zhou, Y.; Zhao, Y. Downregulation of CD9 promotes pancreatic cancer growth and metastasis through upregulation of epidermal growth factor on the cell surface. Oncology Reports 2015, 34, 350–358, doi:10.3892/or.2015.3960.
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96, doi:10.1111/j.1600-0854.2006.00515.x.
- Hemler, M.E. Specific tetraspanin functions. Journal of Cell Biology 2001, 155, 1103–1108, doi:10.1083/jcb.200108061.
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nature Reviews Molecular Cell Biology 2005, 6, 801–811, doi:10.1038/nrm1736.
- Charrin, S.; le Naour, F.; Silvie, O.; Milhiet, P.-E.; Boucheix, C.; Rubinstein, E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 2009, 420, 133–154, doi:10.1042/BJ20082422.
- Hemler, M.E. Integrin associated proteins. Current Opinion in Cell Biology 1998, 10, 578–585, doi:10.1016/S0955-0674(98)80032-X.
- Winterwood, N.E.; Varzavand, A.; Meland, M.N.; Ashman, L.K.; Stipp, C.S. A Critical Role for Tetraspanin CD151 in α3β1 and α6β4 Integrin–dependent Tumor Cell Functions on Laminin-5. Mol Biol Cell 2006, 17, 2707–2721, doi:10.1091/mbc.E05-11-1042.
- Boucheix, C.; Duc, G.H.T.; Jasmin, C.; Rubinstein, E. Tetraspanins and malignancy. Expert Reviews in Molecular Medicine 2001, 3, 1–17, doi:10.1017/S1462399401002381.
- Berditchevski, F. Complexes of tetraspanins with integrins: more than meets the eye. Journal of Cell Science 2001, 114, 4143–4151.
- Yáñez-Mó, M.; Mittelbrunn, M.; Sánchez-Madrid, F. Tetraspanins and Intercellular Interactions. Microcirculation 2001, 8, 153–168, doi:10.1080/mic.8.3.153.168.
- Hemler, M.E. Targeting of tetraspanin proteins — potential benefits and strategies. Nat Rev Drug Discov 2008, 7, 747–758, doi:10.1038/nrd2659.
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010, 73, 1907–1920, doi:10.1016/j.jprot.2010.06.006.
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692, doi:10.1016/j.jmb.2015.09.019.
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241-1244, doi:10.1093/nar/gkr828.
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000, doi:10.1002/pmic.200900351.
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 2019, 47, D516–D519, doi:10.1093/nar/gky1029.
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol 2012, 10, e1001450, doi:10.1371/journal.pbio.1001450.
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288, doi:10.1111/j.1365-2141.1967.tb08741.x.
- Barrachina, M.N.; Calderón‐Cruz, B.; Fernandez‐Rocca, L.; García, Á. Application of Extracellular Vesicles Proteomics to Cardiovascular Disease: Guidelines, Data Analysis, and Future Perspectives. PROTEOMICS 2019, 19, 1800247, doi:10.1002/pmic.201800247.
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688, doi:10.1007/s00018-011-0689-3.
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br J Cancer 1972, 26, 239–257, doi:10.1038/bjc.1972.33.
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593, doi:10.1038/nri2567.
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N Engl J Med 2018, 379, 958–966, doi:10.1056/NEJMra1704286.
- Montoro-García, S.; Shantsila, E.; Marín, F.; Blann, A.; Lip, G.Y.H. Circulating microparticles: new insights into the biochemical basis of microparticle release and activity. Basic Res. Cardiol. 2011, 106, 911–923, doi:10.1007/s00395-011-0198-4.
- Février, B.; Raposo, G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421, doi:10.1016/j.ceb.2004.06.003.
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172, doi:10.1084/jem.183.3.1161.
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705, doi:10.1124/pr.112.005983.
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801, doi:10.1158/0008-5472.CAN-10-4455.
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Nitadori-Hoshino, A.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012, 18, 883–891, doi:10.1038/nm.2753.
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011, 2, 282, doi:10.1038/ncomms1285.
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738, doi:10.3389/fimmu.2018.00738.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228, doi:10.1038/nrm.2017.125.
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: a common pathway for a specialized function. J. Biochem. 2006, 140, 13–21, doi:10.1093/jb/mvj128.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. The Journal of Cell Biology 2013, 200, 373–383, doi:10.1083/jcb.201211138.
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212, doi:10.1016/j.biochi.2006.10.014.
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.-F.; Kobayashi, T.; Salles, J.-P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171, doi:10.1042/BJ20031594.
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Progress in Lipid Research 2017, 66, 30–41, doi:10.1016/j.plipres.2017.03.001.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 2007, 9, 654–659, doi:10.1038/ncb1596.
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. Journal of Oncology 2019, 2019, 1–19, doi:10.1155/2019/1639854.
- Ahmed, K.A.; Xiang, J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mol Med 2011, 15, 1458–1473, doi:10.1111/j.1582-4934.2010.01008.x.
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2015, dmv055, doi:10.1093/humupd/dmv055.
- Zöller, M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nature Reviews Cancer 2009, 9, 40–55, doi:10.1038/nrc2543.
- Rana, S.; Zöller, M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochemical Society Transactions 2011, 39, 559–562, doi:10.1042/BST0390559.
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. The International Journal of Biochemistry & Cell Biology 2012, 44, 1574–1584, doi:10.1016/j.biocel.2012.06.018.
- Kaji, K.; Oda, S.; Shikano, T.; Ohnuki, T.; Uematsu, Y.; Sakagami, J.; Tada, N.; Miyazaki, S.; Kudo, A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet 2000, 24, 279–282, doi:10.1038/73502.
- Le Naour, F.; Rubinstein, E.; Jasmin, C.; Prenant, M.; Boucheix, C. Severely reduced female fertility in CD9-deficient mice. Science 2000, 287, 319–321, doi:10.1126/science.287.5451.319.
- Miyado, K. Requirement of CD9 on the Egg Plasma Membrane for Fertilization. Science 2000, 287, 321–324, doi:10.1126/science.287.5451.321.
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937, doi:10.1111/j.1600-0854.2009.00920.x.
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247, doi:10.1126/science.1153124.
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Developmental Cell 2011, 21, 708–721, doi:10.1016/j.devcel.2011.08.019.
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol 2010, 190, 1079–1091, doi:10.1083/jcb.201002049.
- Bari, R.; Guo, Q.; Xia, B.; Zhang, Y.H.; Giesert, E.E.; Levy, S.; Zheng, J.J.; Zhang, X.A. Tetraspanins regulate the protrusive activities of cell membrane. Biochem. Biophys. Res. Commun. 2011, 415, 619–626, doi:10.1016/j.bbrc.2011.10.121.
- Umeda, R.; Satouh, Y.; Takemoto, M.; Nakada-Nakura, Y.; Liu, K.; Yokoyama, T.; Shirouzu, M.; Iwata, S.; Nomura, N.; Sato, K.; et al. Structural insights into tetraspanin CD9 function. Nat Commun 2020, 11, 1606, doi:10.1038/s41467-020-15459-7.
- Frolikova, M.; Manaskova-Postlerova, P.; Cerny, J.; Jankovicova, J.; Simonik, O.; Pohlova, A.; Secova, P.; Antalikova, J.; Dvorakova-Hortova, K. CD9 and CD81 Interactions and Their Structural Modelling in Sperm Prior to Fertilization. IJMS 2018, 19, 1236, doi:10.3390/ijms19041236.
- Jankovicova, J.; Frolikova, M.; Palenikova, V.; Valaskova, E.; Cerny, J.; Secova, P.; Bartokova, M.; Horovska, L.; Manaskova-Postlerova, P.; Antalikova, J.; et al. Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Scientific Reports 2020, 10, 4374, doi:10.1038/s41598-020-61334-2.
- Sala-Valdés, M.; Ursa, Á.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sánchez-Madrid, F.; Yáñez-Mó, M. EWI-2 and EWI-F Link the Tetraspanin Web to the Actin Cytoskeleton through Their Direct Association with Ezrin-Radixin-Moesin Proteins. J. Biol. Chem. 2006, 281, 19665–19675, doi:10.1074/jbc.M602116200.
- Rana, S.; Claas, C.; Kretz, C.C.; Nazarenko, I.; Zoeller, M. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int. J. Biochem. Cell Biol. 2011, 43, 106–119, doi:10.1016/j.biocel.2010.10.002.
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front Immunol 2014, 5, doi:10.3389/fimmu.2014.00442.
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661, doi:10.1074/jbc.M112.445304.
- Yáñez-Mó, M.; Barreiro, O.; Gordon-Alonso, M.; Sala-Valdés, M.; Sánchez-Madrid, F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009, 19, 434–446, doi:10.1016/j.tcb.2009.06.004.
- Bassani, S.; Cingolani, L.A. Tetraspanins: Interactions and interplay with integrins. The International Journal of Biochemistry & Cell Biology 2012, 44, 703–708, doi:10.1016/j.biocel.2012.01.020.
- Fanaei, M.; Monk, P.N.; Partridge, L.J. The role of tetraspanins in fusion. Biochem. Soc. Trans. 2011, 39, 524–528, doi:10.1042/BST0390524.
- Kolesnikova, T.V.; Kazarov, A.R.; Lemieux, M.E.; Lafleur, M.A.; Kesari, S.; Kung, A.L.; Hemler, M.E. Glioblastoma Inhibition by Cell Surface Immunoglobulin Protein EWI-2, In Vitro and In Vivo. Neoplasia 2009, 11, 77-IN10, doi:10.1593/neo.81180.
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015, 13, 17–24, doi:10.1016/j.gpb.2015.02.001.
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51, doi:10.1016/j.semcdb.2015.02.010.
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles 2014, 3, 24641, doi:10.3402/jev.v3.24641.
- Tian, T.; Zhu, Y.-L.; Hu, F.-H.; Wang, Y.-Y.; Huang, N.-P.; Xiao, Z.-D. Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 2013, 228, 1487–1495, doi:10.1002/jcp.24304.
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005, 5, 136–148, doi:10.1038/nri1548.
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209, doi:10.1016/j.tcb.2008.03.002.
- Johnstone, R.M. Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 2006, 36, 315–321, doi:10.1016/j.bcmd.2005.12.001.
- Lebreton, A.; Séraphin, B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta 2008, 1779, 558–565, doi:10.1016/j.bbagrm.2008.02.003.
- Schorey, J.S.; Bhatnagar, S. Exosome function: from tumor immunology to pathogen biology. Traffic 2008, 9, 871–881, doi:10.1111/j.1600-0854.2008.00734.x.
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678, doi:10.1158/0008-5472.CAN-09-2470.
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266, doi:10.1182/blood-2004-03-0824.
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-exosomes and leukocyte activation: an ambivalent crosstalk. Cell Commun. Signal 2012, 10, 37, doi:10.1186/1478-811X-10-37.
- Stoeck, A.; Keller, S.; Riedle, S.; Sanderson, M.P.; Runz, S.; Le Naour, F.; Gutwein, P.; Ludwig, A.; Rubinstein, E.; Altevogt, P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 2006, 393, 609–618, doi:10.1042/BJ20051013.
- Hawari, F.I.; Rouhani, F.N.; Cui, X.; Yu, Z.-X.; Buckley, C.; Kaler, M.; Levine, S.J. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 1297–1302, doi:10.1073/pnas.0307981100.
- Hakulinen, J.; Junnikkala, S.; Sorsa, T.; Meri, S. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur. J. Immunol. 2004, 34, 2620–2629, doi:10.1002/eji.200424969.
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 17380–17385, doi:10.1073/pnas.1304266110.
- Menck, K.; Klemm, F.; Gross, J.C.; Pukrop, T.; Wenzel, D.; Binder, C. Induction and transport of Wnt 5a during macrophage-induced malignant invasion is mediated by two types of extracellular vesicles. Oncotarget 2013, 4, 2057–2066, doi:10.18632/oncotarget.1336.
- Atay, S.; Gercel-Taylor, C.; Taylor, D.D. Human trophoblast-derived exosomal fibronectin induces pro-inflammatory IL-1β production by macrophages. Am. J. Reprod. Immunol. 2011, 66, 259–269, doi:10.1111/j.1600-0897.2011.00995.x.
- Otto, G.P.; Nichols, B.J. The roles of flotillin microdomains--endocytosis and beyond. J. Cell. Sci. 2011, 124, 3933–3940, doi:10.1242/jcs.092015.
- Volonte, D.; Galbiati, F.; Li, S.; Nishiyama, K.; Okamoto, T.; Lisanti, M.P. Flotillins/cavatellins are differentially expressed in cells and tissues and form a hetero-oligomeric complex with caveolins in vivo. Characterization and epitope-mapping of a novel flotillin-1 monoclonal antibody probe. J. Biol. Chem. 1999, 274, 12702–12709, doi:10.1074/jbc.274.18.12702.
- Bickel, P.E.; Scherer, P.E.; Schnitzer, J.E.; Oh, P.; Lisanti, M.P.; Lodish, H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997, 272, 13793–13802, doi:10.1074/jbc.272.21.13793.
- Frick, M.; Bright, N.A.; Riento, K.; Bray, A.; Merrified, C.; Nichols, B.J. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 2007, 17, 1151–1156, doi:10.1016/j.cub.2007.05.078.
- Jahn, R.; Lang, T.; Südhof, T.C. Membrane fusion. Cell 2003, 112, 519–533, doi:10.1016/s0092-8674(03)00112-0.
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219, doi:10.1371/journal.pone.0005219.
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683, doi:10.1038/nsmb.1455.
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713, doi:10.1007/s00018-016-2346-3.
- Lu, J.; Li, J.; Liu, S.; Wang, T.; Ianni, A.; Bober, E.; Braun, T.; Xiang, R.; Yue, S. Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget 2017, 8, 62803–62815, doi:10.18632/oncotarget.19119.
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016, 126, 1216–1223, doi:10.1172/JCI81136.
- Yue, S.; Mu, W.; Erb, U.; Zöller, M. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2014, 6, 2366–2384, doi:10.18632/oncotarget.2958.
- Mu, W.; Rana, S.; Zöller, M. Host Matrix Modulation by Tumor Exosomes Promotes Motility and Invasiveness. Neoplasia 2013, 15, 875–887.
- Yang, C.; Robbins, P.D. The Roles of Tumor-Derived Exosomes in Cancer Pathogenesis Available online: https://www.hindawi.com/journals/jir/2011/842849/ (accessed on Jul 29, 2020).
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335, doi:10.1038/nature15756.
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459, doi:10.1146/annurev-immunol-041015-055700.
- Szajnik, M.; Czystowska, M.; Szczepanski, M.J.; Mandapathil, M.; Whiteside, T.L. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 2010, 5, e11469, doi:10.1371/journal.pone.0011469.
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600, doi:10.1038/nm0598-594.
- Mittelbrunn, M.; Yáñez-Mó, M.; Sancho, D.; Ursa, A.; Sánchez-Madrid, F. Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J. Immunol. 2002, 169, 6691–6695, doi:10.4049/jimmunol.169.12.6691.
- Rocha-Perugini, V.; Zamai, M.; Gonzalez-Granado, J.M.; Barreiro, O.; Tejera, E.; Yanez-Mo, M.; Caiolfa, V.R.; Sanchez-Madrid, F. CD81 Controls Sustained T Cell Activation Signaling and Defines the Maturation Stages of Cognate Immunological Synapses. Molecular and Cellular Biology 2013, 33, 3644–3658, doi:10.1128/MCB.00302-13.
- Kropshofer, H.; Spindeldreher, S.; Röhn, T.A.; Platania, N.; Grygar, C.; Daniel, N.; Wölpl, A.; Langen, H.; Horejsi, V.; Vogt, A.B. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 2002, 3, 61–68, doi:10.1038/ni750.
- Imai, T.; Kakizaki, M.; Nishimura, M.; Yoshie, O. Molecular analyses of the association of CD4 with two members of the transmembrane 4 superfamily, CD81 and CD82. J. Immunol. 1995, 155, 1229–1239.
- Maecker, H.T.; Levy, S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 1997, 185, 1505–1510, doi:10.1084/jem.185.8.1505.
- Rocha-Perugini, V.; González-Granado, J.M.; Tejera, E.; López-Martín, S.; Yañez-Mó, M.; Sanchez-Madrid, F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol 2014, 44, 1967–1975, doi:10.1002/eji.201344235.
- Unternaehrer, J.J.; Chow, A.; Pypaert, M.; Inaba, K.; Mellman, I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci U S A 2007, 104, 234–239, doi:10.1073/pnas.0609665104.
- Nj, P.; Lk, D.; Pa, R. CDw78 defines MHC class II-peptide complexes that require Ii chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J Immunol 2006, 177, 5451–5458, doi:10.4049/jimmunol.177.8.5451.
- Wright, M.D.; Moseley, G.W.; van Spriel, A.B. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens 2004, 64, 533–542, doi:10.1111/j.1399-0039.2004.00321.x.
- Sheng, K.-C.; Spriel, A.B. van; Gartlan, K.H.; Sofi, M.; Apostolopoulos, V.; Ashman, L.; Wright, M.D. Tetraspanins CD37 and CD151 differentially regulate Ag presentation and T-cell co-stimulation by DC. European Journal of Immunology 2009, 39, 50–55, doi:10.1002/eji.200838798.
References
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. Functional domains in tetraspanin proteins. Trends in Biochemical Sciences 2003, 28, 106–112, doi:10.1016/S0968-0004(02)00014-2.
- Berditchevski, F.; Odintsova, E.; Sawada, S.; Gilbert, E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002, 277, 36991–37000, doi:10.1074/jbc.M205265200.
- Charrin, S.; Manié, S.; Oualid, M.; Billard, M.; Boucheix, C.; Rubinstein, E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002, 516, 139–144, doi:10.1016/s0014-5793(02)02522-x.
- Yang, X.; Claas, C.; Kraeft, S.-K.; Chen, L.B.; Wang, Z.; Kreidberg, J.A.; Hemler, M.E. Palmitoylation of Tetraspanin Proteins: Modulation of CD151 Lateral Interactions, Subcellular Distribution, and Integrin-dependent Cell Morphology. Mol Biol Cell 2002, 13, 767–781, doi:10.1091/mbc.01-05-0275.
- Huang, S.; Yuan, S.; Dong, M.; Su, J.; Yu, C.; Shen, Y.; Xie, X.; Yu, Y.; Yu, X.; Chen, S.; et al. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics 2005, 86, 674–684, doi:10.1016/j.ygeno.2005.08.004.
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205, doi:10.1007/PL00000933.
- Tang, M.; Yin, G.; Wang, F.; Liu, H.; Zhou, S.; Ni, J.; Chen, C.; Zhou, Y.; Zhao, Y. Downregulation of CD9 promotes pancreatic cancer growth and metastasis through upregulation of epidermal growth factor on the cell surface. Oncology Reports 2015, 34, 350–358, doi:10.3892/or.2015.3960.
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96, doi:10.1111/j.1600-0854.2006.00515.x.
- Hemler, M.E. Specific tetraspanin functions. Journal of Cell Biology 2001, 155, 1103–1108, doi:10.1083/jcb.200108061.
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nature Reviews Molecular Cell Biology 2005, 6, 801–811, doi:10.1038/nrm1736.
- Charrin, S.; le Naour, F.; Silvie, O.; Milhiet, P.-E.; Boucheix, C.; Rubinstein, E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 2009, 420, 133–154, doi:10.1042/BJ20082422.
- Hemler, M.E. Integrin associated proteins. Current Opinion in Cell Biology 1998, 10, 578–585, doi:10.1016/S0955-0674(98)80032-X.
- Winterwood, N.E.; Varzavand, A.; Meland, M.N.; Ashman, L.K.; Stipp, C.S. A Critical Role for Tetraspanin CD151 in α3β1 and α6β4 Integrin–dependent Tumor Cell Functions on Laminin-5. Mol Biol Cell 2006, 17, 2707–2721, doi:10.1091/mbc.E05-11-1042.
- Boucheix, C.; Duc, G.H.T.; Jasmin, C.; Rubinstein, E. Tetraspanins and malignancy. Expert Reviews in Molecular Medicine 2001, 3, 1–17, doi:10.1017/S1462399401002381.
- Berditchevski, F. Complexes of tetraspanins with integrins: more than meets the eye. Journal of Cell Science 2001, 114, 4143–4151.
- Yáñez-Mó, M.; Mittelbrunn, M.; Sánchez-Madrid, F. Tetraspanins and Intercellular Interactions. Microcirculation 2001, 8, 153–168, doi:10.1080/mic.8.3.153.168.
- Hemler, M.E. Targeting of tetraspanin proteins — potential benefits and strategies. Nat Rev Drug Discov 2008, 7, 747–758, doi:10.1038/nrd2659.
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010, 73, 1907–1920, doi:10.1016/j.jprot.2010.06.006.
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692, doi:10.1016/j.jmb.2015.09.019.
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241-1244, doi:10.1093/nar/gkr828.
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000, doi:10.1002/pmic.200900351.
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in Extracellular Vesicles. Nucleic Acids Res 2019, 47, D516–D519, doi:10.1093/nar/gky1029.
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol 2012, 10, e1001450, doi:10.1371/journal.pbio.1001450.
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288, doi:10.1111/j.1365-2141.1967.tb08741.x.
- Barrachina, M.N.; Calderón‐Cruz, B.; Fernandez‐Rocca, L.; García, Á. Application of Extracellular Vesicles Proteomics to Cardiovascular Disease: Guidelines, Data Analysis, and Future Perspectives. Proteomics 2019, 19, 1800247, doi:10.1002/pmic.201800247.
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: emerging role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688, doi:10.1007/s00018-011-0689-3.
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br J Cancer 1972, 26, 239–257, doi:10.1038/bjc.1972.33.
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593, doi:10.1038/nri2567.
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N Engl J Med 2018, 379, 958–966, doi:10.1056/NEJMra1704286.
- Montoro-García, S.; Shantsila, E.; Marín, F.; Blann, A.; Lip, G.Y.H. Circulating microparticles: new insights into the biochemical basis of microparticle release and activity. Basic Res. Cardiol. 2011, 106, 911–923, doi:10.1007/s00395-011-0198-4.
- Février, B.; Raposo, G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421, doi:10.1016/j.ceb.2004.06.003.
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172, doi:10.1084/jem.183.3.1161.
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of Extracellular Vesicles. Pharmacol. Rev. 2012, 64, 676–705, doi:10.1124/pr.112.005983.
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by Melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801, doi:10.1158/0008-5472.CAN-10-4455.
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Nitadori-Hoshino, A.; et al. Melanoma Exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012, 18, 883–891, doi:10.1038/nm.2753.
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded Exosomes from T cells to antigen-presenting cells. Nat Commun 2011, 2, 282, doi:10.1038/ncomms1285.
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738, doi:10.3389/fimmu.2018.00738.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228, doi:10.1038/nrm.2017.125.
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: a common pathway for a specialized function. J. Biochem. 2006, 140, 13–21, doi:10.1093/jb/mvj128.
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, microvesicles, and friends. The Journal of Cell Biology 2013, 200, 373–383, doi:10.1083/jcb.201211138.
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212, doi:10.1016/j.biochi.2006.10.014.
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.-F.; Kobayashi, T.; Salles, J.-P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived Exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171, doi:10.1042/BJ20031594.
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in Exosomes: Current knowledge and the way forward. Progress in Lipid Research 2017, 66, 30–41, doi:10.1016/j.plipres.2017.03.001.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 2007, 9, 654–659, doi:10.1038/ncb1596.
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. Journal of Oncology 2019, 2019, 1–19, doi:10.1155/2019/1639854.
- Ahmed, K.A.; Xiang, J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mol Med 2011, 15, 1458–1473, doi:10.1111/j.1582-4934.2010.01008.x.
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular Vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2015, dmv055, doi:10.1093/humupd/dmv055.
- Zöller, M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nature Reviews Cancer 2009, 9, 40–55, doi:10.1038/nrc2543.
- Rana, S.; Zöller, M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochemical Society Transactions 2011, 39, 559–562, doi:10.1042/BST0390559.
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored Exosomes: The exosomal tetraspanin web contributes to target cell selection. The International Journal of Biochemistry & Cell Biology 2012, 44, 1574–1584, doi:10.1016/j.biocel.2012.06.018.
- Kaji, K.; Oda, S.; Shikano, T.; Ohnuki, T.; Uematsu, Y.; Sakagami, J.; Tada, N.; Miyazaki, S.; Kudo, A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet 2000, 24, 279–282, doi:10.1038/73502.
- Le Naour, F.; Rubinstein, E.; Jasmin, C.; Prenant, M.; Boucheix, C. Severely reduced female fertility in CD9-deficient mice. Science 2000, 287, 319–321, doi:10.1126/science.287.5451.319.
- Miyado, K. Requirement of CD9 on the Egg Plasma Membrane for Fertilization. Science 2000, 287, 321–324, doi:10.1126/science.287.5451.321.
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937, doi:10.1111/j.1600-0854.2009.00920.x.
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of Exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247, doi:10.1126/science.1153124.
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Developmental Cell 2011, 21, 708–721, doi:10.1016/j.devcel.2011.08.019.
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol 2010, 190, 1079–1091, doi:10.1083/jcb.201002049.
- Bari, R.; Guo, Q.; Xia, B.; Zhang, Y.H.; Giesert, E.E.; Levy, S.; Zheng, J.J.; Zhang, X.A. Tetraspanins regulate the protrusive activities of cell membrane. Biochem. Biophys. Res. Commun. 2011, 415, 619–626, doi:10.1016/j.bbrc.2011.10.121.
- Umeda, R.; Satouh, Y.; Takemoto, M.; Nakada-Nakura, Y.; Liu, K.; Yokoyama, T.; Shirouzu, M.; Iwata, S.; Nomura, N.; Sato, K.; et al. Structural insights into tetraspanin CD9 function. Nat Commun 2020, 11, 1606, doi:10.1038/s41467-020-15459-7.
- Frolikova, M.; Manaskova-Postlerova, P.; Cerny, J.; Jankovicova, J.; Simonik, O.; Pohlova, A.; Secova, P.; Antalikova, J.; Dvorakova-Hortova, K. CD9 and CD81 Interactions and Their Structural Modelling in Sperm Prior to Fertilization. IJMS 2018, 19, 1236, doi:10.3390/ijms19041236.
- Jankovicova, J.; Frolikova, M.; Palenikova, V.; Valaskova, E.; Cerny, J.; Secova, P.; Bartokova, M.; Horovska, L.; Manaskova-Postlerova, P.; Antalikova, J.; et al. Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Scientific Reports 2020, 10, 4374, doi:10.1038/s41598-020-61334-2.
- Sala-Valdés, M.; Ursa, Á.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sánchez-Madrid, F.; Yáñez-Mó, M. EWI-2 and EWI-F Link the Tetraspanin Web to the Actin Cytoskeleton through Their Direct Association with Ezrin-Radixin-Moesin Proteins. J. Biol. Chem. 2006, 281, 19665–19675, doi:10.1074/jbc.M602116200.
- Rana, S.; Claas, C.; Kretz, C.C.; Nazarenko, I.; Zoeller, M. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int. J. Biochem. Cell Biol. 2011, 43, 106–119, doi:10.1016/j.biocel.2010.10.002.
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front Immunol 2014, 5, doi:10.3389/fimmu.2014.00442.
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward Exosomes. J. Biol. Chem. 2013, 288, 11649–11661, doi:10.1074/jbc.M112.445304.
- Yáñez-Mó, M.; Barreiro, O.; Gordon-Alonso, M.; Sala-Valdés, M.; Sánchez-Madrid, F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009, 19, 434–446, doi:10.1016/j.tcb.2009.06.004.
- Bassani, S.; Cingolani, L.A. Tetraspanins: Interactions and interplay with integrins. The International Journal of Biochemistry & Cell Biology 2012, 44, 703–708, doi:10.1016/j.biocel.2012.01.020.
- Fanaei, M.; Monk, P.N.; Partridge, L.J. The role of tetraspanins in fusion. Biochem. Soc. Trans. 2011, 39, 524–528, doi:10.1042/BST0390524.
- Kolesnikova, T.V.; Kazarov, A.R.; Lemieux, M.E.; Lafleur, M.A.; Kesari, S.; Kung, A.L.; Hemler, M.E. Glioblastoma Inhibition by Cell Surface Immunoglobulin Protein EWI-2, In Vitro and In Vivo. Neoplasia 2009, 11, 77-IN10, doi:10.1593/neo.81180.
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015, 13, 17–24, doi:10.1016/j.gpb.2015.02.001.
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular Vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51, doi:10.1016/j.semcdb.2015.02.010.
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles 2014, 3, 24641, doi:10.3402/jev.v3.24641.
- Tian, T.; Zhu, Y.-L.; Hu, F.-H.; Wang, Y.-Y.; Huang, N.-P.; Xiao, Z.-D. Dynamics of Exosome internalization and trafficking. J. Cell. Physiol. 2013, 228, 1487–1495, doi:10.1002/jcp.24304.
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005, 5, 136–148, doi:10.1038/nri1548.
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant Exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209, doi:10.1016/j.tcb.2008.03.002.
- Johnstone, R.M. Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 2006, 36, 315–321, doi:10.1016/j.bcmd.2005.12.001.
- Lebreton, A.; Séraphin, B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta 2008, 1779, 558–565, doi:10.1016/j.bbagrm.2008.02.003.
- Schorey, J.S.; Bhatnagar, S. Exosome function: from tumor immunology to pathogen biology. Traffic 2008, 9, 871–881, doi:10.1111/j.1600-0854.2008.00734.x.
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of Exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678, doi:10.1158/0008-5472.CAN-09-2470.
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of Exosomes by dendritic cells. Blood 2004, 104, 3257–3266, doi:10.1182/blood-2004-03-0824.
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-Exosomes and leukocyte activation: an ambivalent crosstalk. Cell Commun. Signal 2012, 10, 37, doi:10.1186/1478-811X-10-37.
- Stoeck, A.; Keller, S.; Riedle, S.; Sanderson, M.P.; Runz, S.; Le Naour, F.; Gutwein, P.; Ludwig, A.; Rubinstein, E.; Altevogt, P. A role for Exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 2006, 393, 609–618, doi:10.1042/BJ20051013.
- Hawari, F.I.; Rouhani, F.N.; Cui, X.; Yu, Z.-X.; Buckley, C.; Kaler, M.; Levine, S.J. Release of full-length 55-kDa TNF receptor 1 in Exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 1297–1302, doi:10.1073/pnas.0307981100.
- Hakulinen, J.; Junnikkala, S.; Sorsa, T.; Meri, S. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur. J. Immunol. 2004, 34, 2620–2629, doi:10.1002/eji.200424969.
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer cell Exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 17380–17385, doi:10.1073/pnas.1304266110.
- Menck, K.; Klemm, F.; Gross, J.C.; Pukrop, T.; Wenzel, D.; Binder, C. Induction and transport of Wnt 5a during macrophage-induced malignant invasion is mediated by two types of Extracellular Vesicles. Oncotarget 2013, 4, 2057–2066, doi:10.18632/oncotarget.1336.
- Atay, S.; Gercel-Taylor, C.; Taylor, D.D. Human trophoblast-derived exosomal fibronectin induces pro-inflammatory IL-1β production by macrophages. Am. J. Reprod. Immunol. 2011, 66, 259–269, doi:10.1111/j.1600-0897.2011.00995.x.
- Otto, G.P.; Nichols, B.J. The roles of flotillin microdomains--endocytosis and beyond. J. Cell. Sci. 2011, 124, 3933–3940, doi:10.1242/jcs.092015.
- Volonte, D.; Galbiati, F.; Li, S.; Nishiyama, K.; Okamoto, T.; Lisanti, M.P. Flotillins/cavatellins are differentially expressed in cells and tissues and form a hetero-oligomeric complex with caveolins in vivo. Characterization and epitope-mapping of a novel flotillin-1 monoclonal antibody probe. J. Biol. Chem. 1999, 274, 12702–12709, doi:10.1074/jbc.274.18.12702.
- Bickel, P.E.; Scherer, P.E.; Schnitzer, J.E.; Oh, P.; Lisanti, M.P.; Lodish, H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997, 272, 13793–13802, doi:10.1074/jbc.272.21.13793.
- Frick, M.; Bright, N.A.; Riento, K.; Bray, A.; Merrified, C.; Nichols, B.J. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 2007, 17, 1151–1156, doi:10.1016/j.cub.2007.05.078.
- Jahn, R.; Lang, T.; Südhof, T.C. Membrane fusion. Cell 2003, 112, 519–533, doi:10.1016/s0092-8674(03)00112-0.
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of Exosomes expressing CD63 and caveolin-1 in plasma of Melanoma patients. PLoS ONE 2009, 4, e5219, doi:10.1371/journal.pone.0005219.
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683, doi:10.1038/nsmb.1455.
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: present roles of Extracellular Vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713, doi:10.1007/s00018-016-2346-3.
- Lu, J.; Li, J.; Liu, S.; Wang, T.; Ianni, A.; Bober, E.; Braun, T.; Xiang, R.; Yue, S. Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget 2017, 8, 62803–62815, doi:10.18632/oncotarget.19119.
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016, 126, 1216–1223, doi:10.1172/JCI81136.
- Yue, S.; Mu, W.; Erb, U.; Zöller, M. The tetraspanins CD151 and Tspan8 are essential Exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2014, 6, 2366–2384, doi:10.18632/oncotarget.2958.
- Mu, W.; Rana, S.; Zöller, M. Host Matrix Modulation by Tumor Exosomes Promotes Motility and Invasiveness. Neoplasia 2013, 15, 875–887.
- Yang, C.; Robbins, P.D. The Roles of Tumor-Derived Exosomes in Cancer Pathogenesis Available online: https://www.hindawi.com/journals/jir/2011/842849/ (accessed on Jul 29, 2020).
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335, doi:10.1038/nature15756.
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459, doi:10.1146/annurev-immunol-041015-055700.
- Szajnik, M.; Czystowska, M.; Szczepanski, M.J.; Mandapathil, M.; Whiteside, T.L. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 2010, 5, e11469, doi:10.1371/journal.pone.0011469.
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived Exosomes. Nat. Med. 1998, 4, 594–600, doi:10.1038/nm0598-594.
- Mittelbrunn, M.; Yáñez-Mó, M.; Sancho, D.; Ursa, A.; Sánchez-Madrid, F. Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J. Immunol. 2002, 169, 6691–6695, doi:10.4049/jimmunol.169.12.6691.
- Rocha-Perugini, V.; Zamai, M.; Gonzalez-Granado, J.M.; Barreiro, O.; Tejera, E.; Yanez-Mo, M.; Caiolfa, V.R.; Sanchez-Madrid, F. CD81 Controls Sustained T Cell Activation Signaling and Defines the Maturation Stages of Cognate Immunological Synapses. Molecular and Cellular Biology 2013, 33, 3644–3658, doi:10.1128/MCB.00302-13.
- Kropshofer, H.; Spindeldreher, S.; Röhn, T.A.; Platania, N.; Grygar, C.; Daniel, N.; Wölpl, A.; Langen, H.; Horejsi, V.; Vogt, A.B. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 2002, 3, 61–68, doi:10.1038/ni750.
- Imai, T.; Kakizaki, M.; Nishimura, M.; Yoshie, O. Molecular analyses of the association of CD4 with two members of the transmembrane 4 superfamily, CD81 and CD82. J. Immunol. 1995, 155, 1229–1239.
- Maecker, H.T.; Levy, S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 1997, 185, 1505–1510, doi:10.1084/jem.185.8.1505.
- Rocha-Perugini, V.; González-Granado, J.M.; Tejera, E.; López-Martín, S.; Yañez-Mó, M.; Sanchez-Madrid, F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol 2014, 44, 1967–1975, doi:10.1002/eji.201344235.
- Unternaehrer, J.J.; Chow, A.; Pypaert, M.; Inaba, K.; Mellman, I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci U S A 2007, 104, 234–239, doi:10.1073/pnas.0609665104.
- Nj, P.; Lk, D.; Pa, R. CDw78 defines MHC class II-peptide complexes that require Ii chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J Immunol 2006, 177, 5451–5458, doi:10.4049/jimmunol.177.8.5451.
- Wright, M.D.; Moseley, G.W.; van Spriel, A.B. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens 2004, 64, 533–542, doi:10.1111/j.1399-0039.2004.00321.x.
- Sheng, K.-C.; Spriel, A.B. van; Gartlan, K.H.; Sofi, M.; Apostolopoulos, V.; Ashman, L.; Wright, M.D. Tetraspanins CD37 and CD151 differentially regulate Ag presentation and T-cell co-stimulation by DC. European Journal of Immunology 2009, 39, 50–55, doi:10.1002/eji.200838798.