
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Clodia Osipo | + 726 word(s) | 726 | 2020-10-09 08:41:01 | | | |
| 2 | Nicole Yin | Meta information modification | 726 | 2020-10-21 09:54:24 | | |
Video Upload Options
Roles of Notch signaling in human development and cancer are reviewed herein. The four Notch paralogs along with the five Notch ligands are described. Their structures, mode of activation, and functions are briefly summarized based on published works.
1. Introduction
Notch signaling is an evolutionary conserved pathway, originally discovered through investigations of Drosophila wing development[1] and has since grown into an increasingly large field of study for cancer biologists. This intricate pathway mediates normal stem cell differentiation, cell fate, and organ development[2][3]. However, its dysregulation and role in promoting cellular transformation has led to further investigations of the role of Notch in a variety of cancers[4].
2. Notch Receptors
There exist four known mammalian Notch receptors, Notch1, Notch2, Notch3, and Notch4. Each receptor is translated as a single polypeptide that is subsequently cleaved in the Golgi-apparatus by a furin-like convertase. The resulting cleaved protein is delivered to the plasma membrane as a heterodimeric protein containing an extracellular domain tethered to the transmembrane and intracellular domains by a calcium cation (Figure 1). Upon interaction of the extracellular domain with one of its ligands that include Jagged-1 (JAG1), Jagged-2 (JAG2), Delta-like 1 (DLL1), Delta-like 3 (DLL3), or Delta-like 4 (DLL4), through cell-to-cell contact (Figures 1 and 2), the extracellular portion of the receptor is pulled away from the transmembrane/intracellular domains by ligand-mediated endocytosis. The remaining transmembrane portion of the receptor (NotchTM) is first cleaved by a disintegrin and metalloprotease (ADAM17 or ADAM10), resulting in a product: Notch extracellular truncation (NEXT). NEXT is subsequently cleaved by the γ-secretase complex releasing the intracellular portion of Notch (NotchIC). NotchIC is translocated from the cytoplasm to the nucleus where it binds to the CSL (CBF-1/RBPJ-κ in Homo sapiens/Mus musculus, respectively, Suppressor of Hairless in Drosophila melanogaster, Lag-1 in Caenorhabditis elegans) transcription factor. The interaction of NotchIC with CSL replaces corepressors with coactivators including the transcriptional activator Mastermind1 (MAML1) at regulatory sequences of gene targets (Figure 2). This allows for transcriptional activation of Notch target genes[5][6].
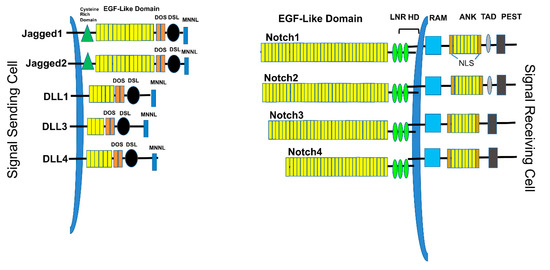
Figure 1. Notch ligands and receptors. The Signal-sending cell expresses Notch ligands, type I transmembrane proteins. These include Jagged1, Jagged2, DLL1, DLL3, and DLL4. Each ligand contains a short cytoplasmic tail, followed by the transmembrane domain, EGF-like repeats, a DOS domain, DLS binding domain, and MNNL domain. The signal-receiving cells express the Notch receptors, including Notch1, Notch2, Notch3, and Notch4. Each receptor contains EGF-like domains, LNR (Lin-Notch repeats), HD (heterodimerization domain), transmembrane domain, RAM (RBPJ-associated molecule), ANK (Ankyrin repeats), TAD (transactivation domain), and PEST (Pro-Glu-Ser-Thr) domains.
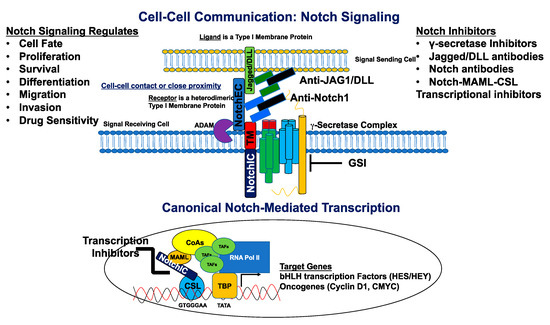
Figure 2. Canonical Notch signaling and inhibitors. The signal-sending cell expressing a Notch ligand (Jagged/DLL) is in close proximity to the signal-receiving cell expressing a Notch receptor. The ligand binds the Notch receptor through EGF-like repeats, and this interaction dislodges the extracellular portion of Notch (NotchEC) from the transmembrane portion (NotchIC-TM). NotchEC is endocytosed with the ligand into the signal sending cell. The removal of NotchEC exposes the ADAM cleavage site on the ecto-domain of the NotchIC-TM. ADAM-mediated cleavage creates a substrate (NEXT: Notch extracellular truncation) for the γ-secretase complex. This subsequent cleavage produces a product (NotchIC) that translocates to the nucleus. NotchIC binds to CSL and recruits MAML. MAML recruits other transcriptional co-activators and the RNA POL II initiation machinery to initiate transcription of Notch target genes including basic helix loop helix (bHLH) proteins of the HES and HEY family of transcriptional repressors. Other genes include the oncogenes, CMYC and CCND1 (Cyclin D1) for initiation of the cell cycle. Notch signaling is inhibited by a variety of molecules that include γ-secretase inhibitors (GSIs), antibodies directed against Notch ligands and receptors, and transcriptional inhibitors that target the NotchIC-MAML-CSL ternary complex. Notch regulates cell fate, proliferation, survival, differentiation, migration, invasion, and sensitivity to cancer drugs.
Some of the earliest known targets of Notch signaling include transcriptional repressors, such as the hairy/enhancer of split (HES) genes, as well as the HES subfamily members HEY1, HEY2, and HEYL[7][8]. These HES/HEY genes are critical cell-fate regulators during development and tissue renewal. In addition to this, cell-cycle regulators such as c-Myc[9] and cyclin D1[10] are directly activated by Notch signaling. Dysregulation of Notch signaling, such as activating Notch receptor mutations, overexpression of ligands and/or receptors, and/or overexpression of its target genes, contributes to increased proliferation, cell transformation, and increased drug resistance in cancers of the breast, multiple myeloma, prostate, T-cell acute lymphoblastic leukemia, and others[11].
References
- S Artavanis-Tsakonas; K Matsuno; M E Fortini; Notch signaling. Science 1995, 268, 225-232, 10.1126/science.7716513.
- Lai, E.C. Notch signaling: Control of cell communication and cell fate. Development 2004, 131, 965–973.
- Chiba, S. Concise review: Notch signaling in stem cell systems. Stem Cells 2006, 24, 2437–2447.
- Lucio Miele; Ingrid Espinoza; Radhika Pochampally; Kounosuke Watabe; Fei Xing; Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. OncoTargets and Therapy 2013, 6, 1249-1259, 10.2147/ott.s36162.
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612.
- Kopan, R.; Ilagan, M.X.G.; Ilagan, M.X.G. The canonical notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233.
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646.
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004, 18, 901–911.
- Apostolos Klinakis; Matthias Szabolcs; Katerina Politi; Hippokratis Kiaris; Spyros Artavanis-Tsakonas; Argiris Efstratiadis; Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proceedings of the National Academy of Sciences 2006, 103, 9262-9267, 10.1073/pnas.0603371103.
- Brenda Cohen; Mamiko Shimizu; Julia Izrailit; Nancy F. L. Ng; Yuri Buchman; James G. Pan; Judy Dering; Michael Reedijk; Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Research and Treatment 2009, 123, 113-124, 10.1007/s10549-009-0621-9.
- Victoria Bolós; Joaquín Grego-Bessa; José Luis De La Pompa; Notch Signaling in Development and Cancer. Endocrine Reviews 2007, 28, 339-363, 10.1210/er.2006-0046.




