| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Danijela Poljuha | + 1012 word(s) | 1012 | 2020-10-12 09:42:10 | | | |
| 2 | Catherine Yang | + 2 word(s) | 1014 | 2020-10-21 08:17:40 | | |
Video Upload Options
Some wild, morphologically diverse taxa of the genus Iris L. in the broad Alpine-Dinaric area have never been explored molecularly, and/or have ambiguous systematic status. The main aims of our research were to perform a molecular study of critical Iris taxa from that area (especially a narrow endemic species I. adriatica, for which we also analysed genome size) and to explore the contribution of eight microsatellites and highly variable chloroplast DNA (ndhJ, rpoC1) markers to the understanding of the Iris taxa taxonomy and phylogeny.
1. Introduction
Iris L. (family Iridaceae) is a diverse genus with over 300 taxa distributed worldwide, mostly in the northern hemisphere [1], and cultivated elsewhere [2]. Most of the European native taxa of the genus belong to the subgenus Iris L., section Iris L. (“Pogoniris”), and less prevalent are taxa from the subgenus Limniris (Tausch) Spach, section Limniris (Tausch) Spach (“Apogoniris”). Their variety has resulted in ambiguous systematic status of some regional, especially endemic, Iris taxa, often recognised in the national and regional floras [3][4], but with an unclear phylogenetic and classification status. Some of them neither are accepted in the World Checklist of Selected Plant Families [5] nor are molecularly researched in detail. Therefore we intended to molecularly study some, insufficiently researched and/or globally neglected taxa from that area: I. x croatica Horvat et M. D. Horvat, I. illyrica Tomm. ex Vis., I. sibirica L. subsp. erirrhiza (Posp.) Wraber and I. x rotschildii Degen. Furthermore, we paid special attention to the validly described [6] and accepted [5], molecularly unexplored endemic species I. adriatica Trinajstić ex Mitić (Figure 1), a narrow endemic dwarf plant, confined to a few Croatian localities in the wider area of Dalmatia (Figure 2) and classified as a NT (near threatened) species [4].
Figure 1. Narrow endemic wild Alpine-Dinaric endemic species Iris adriatica: (a–c) Individuals of different colours (Photo: Miroslav Mitić).
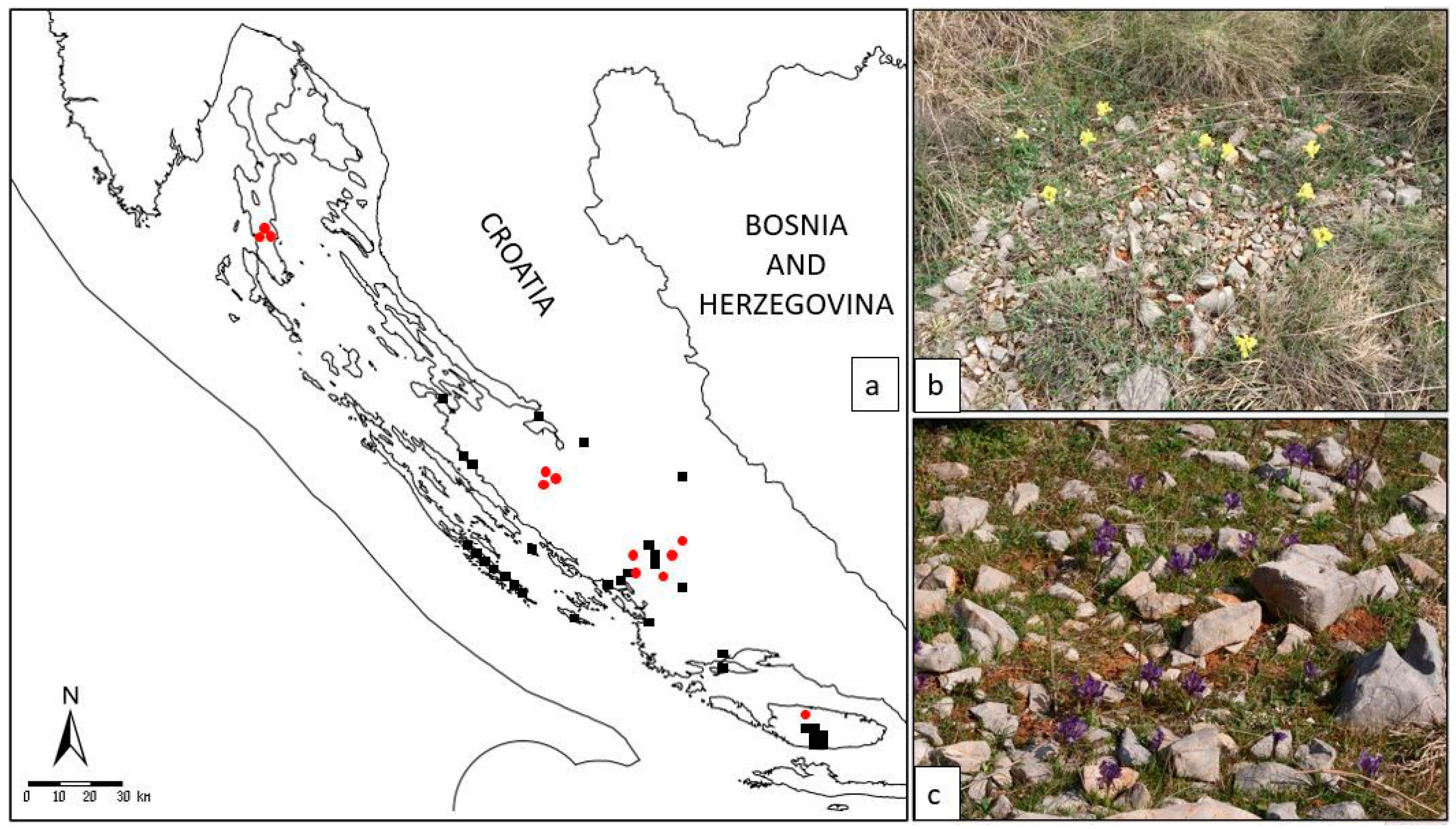
Figure 2. (a) Distribution map of the narrow endemic Alpine-Dinaric species Iris adriatica included in our study (all localities are in Croatia, and are incorporated in the national Flora Croatica Database (https://hirc.botanic.hr/fcd)—FCD; marks: —earlier data from the FCD; —localities of collected specimens in our study); (b) habitat on the locality Brnjica-Pokrovnik; (c) habitat on the island of Cres.
2. Molecular study of selected taxonomically critical taxa of the genus Iris L. from the broader Alpine-Dinaric area
To provide molecular insights into phylogenetic relationships of selected wild Iris taxa of the wider Alpine-Dinaric area (including the adjacent areas of Mediterranean and the Pannonian Plain), with a special emphasis on regional endemics and molecular evidence for their conservation, the aims of our research were: (i) To characterise representative and critical Iris taxa from the wider Alpine-Dinaric area by nuclear (SSR) markers; (ii) to clarify the genetic divergence within and between several wild (local endemic) and cultivated Iris populations through chloroplast DNA (cpDNA) markers; (iii) to present the first molecular description of a nearly threatened narrow endemic dwarf species I. adriatica; and (iv) contribute to the efforts of establishing optimal molecular markers for detecting taxonomic and phylogenetic relationships within critical taxa of the genus Iris.
In total, 32 Iris samples across the wider Alpine-Dinaric region were analysed. We applied 8 SSR markers (IM93, IM123, IM164, IM196, IM200, IM327, IM348, IM391) developed by Tang et al. [7] which proved to be highly polymorphic and amplified alleles across the 39 Iris ecotypes and cultivars. We also applied the maximum likelihood (ML) analysis in reconstructing phylogenetic relationships of a heterogeneous group of Iris species based on two plastid markers - rpoC1 and ndhJ, proposed as good candidate markers for plants barcoding [8][9], and previously not tested in any Iris genus study.
Our results are mostly in agreement with previous studies and monographs of the genus Iris [1][10]. Both the microsatellite-based UPGMA and plastid markers-based maximum likelihood analysis discriminated three main clusters in the set of 32 analysed samples. Two of three clusters covered taxa from the subgenus Iris, section Iris (“Pogoniris”), while the taxa from the subgenus Limniris, section Limniris (“Apogoniris”) were grouped in the third cluster. Our results correspond well to the lower taxonomic categories of the genus, and support separate status of ambiguous regional taxa (e.g., I. sibirica subsp. erirrhiza, I. x croatica and I. x rotschildii).
The first molecular data on I. adriatica revealed its genome size (2C = 12.639 ± 0.202 pg) pointing to relationship with other dwarf irises from the I. pumila complex. We also documented diversity of different populations of the species I. adriatica, showing the existence of geographical ecotypes.
Only a few SSR markers were needed to identify (distinguish) ecotypes and species, while chloroplast markers ndhJ and rpoC1 provided a weaker resolution into the species. However, analysis of sequence data is quicker and much less prone to human error. Chloroplast markers can give further context to SSR analysis and provide independent control despite their lower resolution as they can confirm broader clusters.
3. Conclusions
In the present molecular study of selected representative and critical Iris taxa from the wider Alpine-Dinaric area, we enhanced the current knowledge and understanding of the genus Iris taxonomy and phylogeny. The first molecular data on the nearly threatened narrow endemic dwarf species I. adriatica are particularly important for its further protection and conservation. Our research showed taxonomic positions of investigated taxa within the genus Iris, which is mostly in accordance with previous comprehension of the genus Iris. Additionally, we stressed some, presently unresolved, key taxonomic and phylogenetic questions about certain critical groups and/or taxa of the genus Iris from that area.
Regarding the contribution to the efforts of establishing optimal molecular markers for detecting taxonomic and phylogenetic relationships within critical taxa of the genus Iris, we would recommend the utilisation of SSR markers for subsequent analysis supplemented with a combination of plastid markers until a plastid marker combination for the genus is established and fully validated as convention. For future studies of the genus Iris we would additionally recommend the inclusion of other appropriate barcoding regions to serve the same purpose and hopefully increase the sequencing resolution.
Molecular evidences obtained in this study, besides contribution to the knowledge on taxonomy and phylogeny of the genus Iris in the Alpine-Dinaric, Mediterranean and Pannonian area, should also help in further understanding about the importance of wild, especially endemic Iris taxa and encourage their more intensive conservation efforts.
The article has been published on https://doi.org/10.3390/plants9091229
References
- Mathew, B. The Iris. BT Batsford Ltd, London, UK, 1989.
- Köhlein, F. Iris. Verlag Eugen Ulmer, Stuttgart, Germany, 1981.
- Wraber, T. Iris. In Mala flora Slovenije: ključ za določanje praprotnic in semenk, 3rd ed.; Martinčić, A.; Wraber, T.; Jogan, N.; Ravnik, V.; Podobnik, A.; Turk, B.; Vreš, B., Eds.; Tehniška založba Slovenije, Ljubljana, Slovenia, 1999, pp. 657-659
- Nikolić T, Ed. Flora Croatica Database. Available online: http://hirc.botanic.hr/fcd. Croatia: Department of Biology, Faculty of Science, University of Zagreb.[Accessed 2020 March 29].
- WCSP - World Checklist of Selected Plant Families. Facilitated by the Royal Botanic Gardens, Kew. Available online: http://wcsp.science.kew.org (Accessed on 27 May 2020).
- Mitić, B. Iris adriatica (Iridaceae), a new species from Dalmatia (Croatia). Phyton 2002, 42(2), 305-314.
- Tang, S.; Okah, R.A.; Cordonnier-Pratt, M.M.; Pratt, L.H.; Johnson, V.E.; Taylor, C.A.; Arnold, M.L.; Knapp, S.J. EST and EST-SSR marker resources for Iris. BMC Pl. Biol. 2009, 9(1), 72, DOI:10.1186/1471-2229-9-72.
- Saddhe, A.A.; Kumar, K. DNA barcoding of plants: Selection of core markers for taxonomic groups. Plant Sci. Today 2017, 5, 9–13.
- Blazier, J.C.; Jansen, R.K.; Mower, J.P.; Govindu, M.; Zhang, J.; Weng, M.; Ruhlman, T.A. Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 2016, 117, 1209–1220.
- Wilson, C.A. Subgeneric classification in Iris re-examined using chloroplast sequence data. Taxon 2011, 60(1), 27–35, DOI:10.1002/tax.601004.