
Video Upload Options
The small nucleolar RNA snR30 (U17 in humans) plays a unique role during ribosome synthesis. Unlike most members of the H/ACA class of guide RNAs, the small nucleolar ribonucleoprotein (snoRNP) complex assembled on snR30 does not direct pseudouridylation of ribosomal RNA (rRNA), but instead snR30 is critical for 18S rRNA processing during formation of the small subunit (SSU) of the ribosome. Specifically, snR30 is essential for three pre-rRNA cleavages at the A0/01, A1/1, and A2/2a sites in yeast and humans, respectively. Accordingly, snR30 is the only essential H/ACA guide RNA in yeast. However, the molecular mechanism of snR30 and how it promotes pre-rRNA processing remains under investigation.
1. Overview
Eukaryotic ribosomes are synthesized in the nucleolus with the help of hundreds of assembly factors. In a highly orchestrated fashion, multiple protein and RNA assembly factors are recruited to nascent pre-ribosomal RNA (pre-rRNA) and depart at defined times, whereas ribosomal proteins also associate in a defined order, eventually forming ribosomal subunits together with the matured rRNAs. Among the ribosome assembly factors, the nucleolar snR30 is an essential H/ACA RNA that is critical for the formation of the small ribosomal subunit, but only limited information is available on the mechanistic role of snR30 during the complex process of ribosome formation [1].
2. Introduction
Ribosome biogenesis occurs co-transcriptionally on the pre-rRNAs, and snR30 is also thought to act co-transcriptionally during the processing of 18S rRNA. Three of the four mature rRNAs (18S, 5.8S, and 25S rRNA) are transcribed by RNA polymerase I as a single 35S precursor rRNA (pre-rRNA, 47S pre-rRNA in humans), whereas the fourth rRNA, 5S rRNA, is transcribed independently by RNA Polymerase III. In addition to the mature rRNA sequences, the 35S pre-rRNA contains two external and two internal transcribed spacers abbreviated as ETS and ITS respectively, which are removed through several nucleolytic cleavage events such that these regions are not present in the mature ribosome. The initial site of pre-rRNA cleavage, that separates the 18S rRNA from the remaining pre-rRNA, are called A sites, specifically A0, A1, A2, and A3 in yeast and 01, 1, and 2 in humans (Figure 1) [2][3][4]. Notably, snR30 is involved in pre-rRNA cleavage at sites A0, A1, and A2.
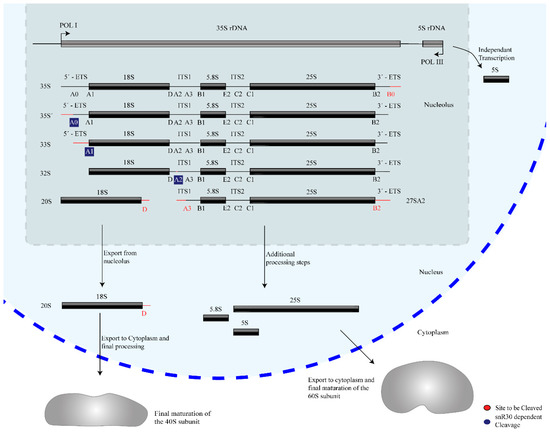
Figure 1. Overview of rRNA processing in yeast. The mature 18S, 5.8S, and 25S rRNAs are generated from a 35S pre-rRNA precursor through several nucleolytic cleavage events, shown here generating characteristic pre-rRNA intermediates. For each step, the subsequent site(s) of cleavage is highlighted in red. snR30 is essential for cleavage at sites A0, A1, and A2, which are highlighted in blue.
During ribosome biogenesis, snR30 does not act alone, but rather forms a stable ribonucleoprotein (RNP) that belongs to the family of H/ACA small nucleolar RNPs (snoRNPs). The snoRNPs represent a large portion of the ribosome biogenesis factors found in the nucleolus, and most snoRNPs chemically modify rRNA, but notably, this is not the case for the snR30 snoRNP. Occurring in the form of either C/D box or H/ACA box guide RNA systems, snoRNPs direct site-specific 2′-O-methylation and pseudouridylation, respectively [8][9]. Each of these complexes is composed of one guide RNA, also called small nucleolar RNA (snoRNA), and four core proteins. The proteins Nop1 (Fibrillarin in vertebrates), Nop56, Nop58, and Snu13 (15.5K) assemble on C/D box guide RNAs, forming C/D snoRNPs [10][11][12][13], whereas the proteins Cbf5 (Dyskerin), Nop10, Gar1, and Nhp2 are components of the H/ACA box snoRNP together with H/ACA box guide RNA [14][15][16]. During ribosome assembly, the H/ACA snoRNPs introduce many pseudouridines in rRNA which play an important role in the maintenance of ribosome structure, stability, and translational fidelity [17][18][19][20][21][22][23].
Interestingly, of the more than one hundred snoRNAs known in yeast, only three guide RNAs are essential, namely U3, U14, and snR30. In addition, deletion of S. cerevisiae snR10 results in slow growth, and a cold-sensitive phenotype [24]. Remarkably, the primary function of these essential snoRNAs is not rRNA modification, but they are all directly or indirectly required for rRNA processing during ribosome synthesis. Additionally, snR30 may also play a role in cholesterol trafficking in higher eukaryotes [25][26][27]. In comparison to snR30, much more is known about the essential U3 snoRNA, a C/D box RNA that is responsible for the correct folding of the central pseudoknot in the 18S rRNA [28]. The central pseudoknot is an SSU rRNA structure conserved from prokaryotes to higher eukaryotes that connects the different domains of the SSU rRNA [29]. To facilitate pseudoknot formation, U3 makes multiple essential base pair interactions to the pre-rRNA in both the 5′ ETS and the 18S rRNA [28]. In particular, the box A and A′ motifs in the U3 snoRNA bind to the 18S rRNA regions which form the central pseudoknot. When U3 is deleted, biogenesis of the small subunit is halted early due to the U3 snoRNP having a central role in biogenesis. This leads to accumulation of 23S pre-rRNA due to an inability to process at A0, A1, or A2 [30]. Lastly, U14 is another C/D RNA that is essential for 18S rRNA formation [31]. U14 appears to have two distinct roles, as one region, that base pairs to the pre-rRNA and is not essential, guides the introduction of a 2′-O-methylation at C414 in 18S rRNA, whereas another region is essential and binds to the 18S rRNA extensively in the 5′ domain of the 18S rRNA [32][33][34]. Depletion of U14’s essential region leads to the exact same phenotype as depletion of U3 or snR30. Thereby, U14 resembles the H/ACA box RNA snR10 which also has dual functions by directing pseudouridine formation at position 2923 in 25S rRNA and by contributing to 18S rRNA processing. In cases where snR10 is lacking, the phenotype is similar to a knockout of the essential genes, except less pronounced. There is minor 35S pre-rRNA accumulation that leads to an increase in 23S pre-rRNA and a 21S pre-rRNA product. However, at permissive temperatures, this pre-rRNA is still processed, albeit slowly, into mature 18S rRNA [35].
Knowledge of ribosome biogenesis, and specifically small subunit biogenesis, has significantly increased due to recent advances in cryo-electron microscopy (cryo-EM), yielding high-resolution structures of the SSU processome complex [36][37][38], but no structural insight of snR30 bound to the SSU processome is available to date. The recent cryo-EM structures provide insight into the interactions within the SSU processome which has led to advances in both the understanding of protein positioning and timings of large-scale rearrangements in the small ribosomal subunit during ribosome biogenesis. However, many of the early-acting factors, such as the snR30 snoRNP, are not present in the structures solved so far, presumably because the snR30 snoRNP has already dissociated before formation of the stable SSU processome. Therefore, no structural information is currently available on the interaction of snR30 with the SSU processome, but instead, our knowledge is derived mostly from genetic and biochemical studies. For example, by tagging and purifying a known biogenesis factor, the timing of SSU processome association of this factor can be roughly estimated by identifying other co-purified factors [39][40]. More recently, different ribosomal precursors have also been studied by expressing and purifying truncated and tagged pre-rRNA variants followed by proteomic analysis, providing additional information on when assembly factors associate and dissociate during pre-rRNA transcription [41][42][43][44]. Notably, taking such an approach, snR30 was detected in early ribosomal intermediates that arise before 18S rRNA transcription is complete, but it was no longer detected in later intermediates [41]. These findings suggest that snR30 acts relatively early during ribosome biogenesis when the structure of the SSU processome might still be rather flexible and labile.
The early stages of eukaryotic ribosome synthesis are characterized by a dynamic interaction network of several factors, including snR30. Ribosome biogenesis starts with the binding of the UTP-A complex to the 5′ ETS of the 35S pre-rRNA transcript. Subsequently, the U3 snRNP, UTP-B, and Mpp10 complexes associate [41][42][45]. While these complexes contain a large portion of the assembly factors, there are many individual proteins, small complexes, and modification enzymes which also act during this period. snR30 is part of one of these complexes that seems to only be present until the 3′ minor domain of 18S rRNA is transcribed [41]. The primary interaction point of snR30 is within expansion segment 6 (ES6) in the central domain, which is unstructured and therefore not resolved in the cryo-EM structures of the SSU processome [38]. The possible function of snR30 binding for pre-rRNA folding will be further discussed below.
By recruiting all these assembly factors, the pre-rRNA in the SSU processome is held in a state where the mature subunit is recognizable, but not fully formed, as evident in the cryo-EM structures [36][37][38], and snR30 may contribute to preventing premature interactions as further discussed below. One of the most striking differences between the SSU processome structure compared to the mature 40S subunit is that the central pseudoknot is unable to form due to the presence of Sas10, Lcp5, the U3 snRNP, and other factors [37]. Furthermore, the SSU processome separates the four domains of the 18S rRNA into independent units which can fold separately [38][46]. More recent structural information from the thermophilic fungus Chaetomium thermophilum has suggested that the 5′, central, 3′ major, and 3′ minor domains of 18S rRNA are not sequentially integrated into the pre-40S subunit, but that the 5′ domain may join the SSU processome last; however, it remains to be investigated whether this finding holds true in other organisms [47]. Thus, the early stages of SSU formation are characterized by dynamic interactions and several conformational changes in rRNA which are facilitated by many factors, including snR30.
In summary, research into ribosome biogenesis is a rapidly evolving field as the concerted functions of hundreds of poorly understood factors and steps including snR30 need to be identified.
3. snR30 Biogenesis, Sequence, and Structure
3.1. Transcription and Processing
In unicellular eukaryotes, such as S. cerevisiae, snR30 is transcribed as an independent gene by RNA Pol II, the mRNA producing polymerase. Contrary to mRNA, the snoRNA undergoes processing to remove the polyA tail by initially binding the Nrd1-Nab3-Sen1 complex which presents the snR30 3′ end to the exosome for exonucleolytic processing [48][49]. On the 5′ end, snR30 possesses a trimethyl guanosine cap like most other H/ACA snoRNAs in yeast [25][50], which is added by the conserved methyltransferase Tgs1 [51].
In multicellular eukaryotes such as humans, transcription and processing of U17a and U17b, two human homologs of snR30, is significantly different. Both U17a and U17b are encoded by the U17HG gene upstream of the cell cycle regulatory gene RCC1 [52]. Interestingly, the U17HG gene harbors U17a and U17b in two introns; however, the U17HG gene seems not to encode a protein and only the U17 sequences are conserved [53]. However, in other species, the location of the U17 gene varies such as in X. laevis, where U17 is transcribed in all six introns of the r-protein S8 gene [54]. Following transcription of the host gene, the excised intron containing U17 is then processed exonucleolytically at both the 5′ and 3′ ends in both Xenopus and HeLa cells, implying a conserved mode of maturation [55][56]. The 3′ end is processed by the exosome [57], whereas the 5′ end is processed by an unknown endonuclease [55][58]. Further information on snoRNA biogenesis and processing events have been reviewed by Kufel and Grzechnik [59].
3.2. snR30 snoRNP Formation and Nucleolar Localization
In yeast, the core snR30 RNP comprising the RNA and the four canonical H/ACA proteins is formed in the same way as other H/ACA snoRNPs that modify rRNA. This process begins with formation of a protein complex initiated by the binding of the protein Shq1 to Cbf5 in the cytoplasm [60]. Shq1 associates with the RNA-binding domain of Cbf5 mimicking interactions of H/ACA snoRNA, which is known to be strongly bound by Cbf5 [61][62]. The Shq1-Cbf5 complex is then shuttled into the nucleus where the majority of RNP maturation takes place. During this process, Nop10, Nhp2, and an assembly factor called Naf1 bind, forming a large protein complex [63]. At the site of snR30 transcription, a pair of ATPases catalyzes the release of Cbf5 from Shq1, freeing the enzyme to tightly bind the snoRNA in its place [64]. This interaction is mediated by Naf1 binding to the large subunit of RNA polymerase II [65]. After binding of Cbf5 to the H/ACA guide RNA, the subsequent maturation step involves the competitive binding of Gar1 to Cbf5, resulting in the displacement of Naf1 which is recycled back to the cytoplasm for further maturation. The now fully assembled H/ACA snoRNP is shuttled from Cajal bodies to the nucleolus by Nopp140 [63][66]. Localization signals for transport into the nucleolus appear to be located in the H and ACA boxes, as well as the general structure of the guide RNA, at least for vertebrates [67][68]. For U17, the ACA box is required for assembly in vitro and presumably in vivo, signifying that nucleolar localization is dependent upon formation of the RNP [69]. As only mature snoRNPs are found within the nucleolus, maturation must occur between transcription in the nucleoplasm and import into the nucleolus [70]. It is possible that some steps of snR30 maturation may occur within Cajal bodies similar to the maturation of other snoRNAs. Immature C/D box RNPs as well as a subgroup of H/ACA RNAs called H/ACA small Cajal body RNAs (scaRNA) can be readily detected within Cajal bodies, and some evidence suggests that H/ACA snoRNPs may in general also traverse Cajal bodies [71][72][73]. In conclusion, current evidence suggests that the maturation of the snR30/U17 RNP follows the same steps as canonical H/ACA guide RNA as all H/ACA RNAs including snR30 assemble with the same proteins.
3.3. Conservation of snR30/U17 Structure, Sequence, and Motifs
Typical H/ACA guide RNAs in eukaryotes share a similar secondary structure comprised of two hairpins connected by a hinge region. The two hairpins are followed by two conserved sequence motifs, the H Box (ANANNA) and the ACA Box, respectively. While most known H/ACA RNAs contain two hairpins in eukaryotes, there are instances of H/ACA guide RNAs having one or three hairpins in selected eukaryotic organisms as well as in archaea. snR30 is an unusual H/ACA guide RNA that has two primary hairpins, the 5′ and 3′ hairpins, but also possesses a third internal hairpin as well as a 41-nt leader sequence at its 5′ end (Figure 2). Notably, the 5′ hairpin of snR30 is much longer than a standard H/ACA hairpin such that S. cerevisiae snR30 has an unusual length of 606 nt, almost triple the length of the average yeast H/ACA guide RNA (~200 nt). While not found in all H/ACA snoRNAs, the internal hairpin and the 41-nt leader are both features that are also present in some other H/ACA guide RNAs [74]. Interestingly, snR30 lacks an unpaired internal bulge following the first stem of the 5′ hairpin, a feature known as the pseudouridylation pocket in standard H/ACA guide RNAs. In contrast, the 3′ hairpin contains a single-stranded bulge like all other H/ACA guide RNA hairpins, and the top of the bulge is located at a conserved 14–16 nucleotide distance from the base of the hairpin and the ACA box [75]. In H/ACA snoRNAs directing pseudouridylation, this distance is important for properly positioning the guide RNA on the Cbf5-Nop10-Nhp2 binding surface, allowing binding of the target RNA to the pseudouridylation pocket and positioning of the target uridine into the active site of Cbf5 [62][75]. In the 5′ hairpin of snR30, the only similar bulge occurs too far away from the base of the stem and the H box for correct positioning of Cbf5. Accordingly, no pseudouridine has been suggested to be introduced by the 5′ hairpin of snR30. Similar to canonical H/ACA snoRNAs and based on the location of the H and ACA Boxes, snR30 is expected to bind one set of the H/ACA core proteins (Cbf5, Nop10, Gar1, and Nhp2) to each of the 5′ and 3′ hairpins, resulting in a predicted 2:1 stoichiometry between the proteins and the snR30 RNA. Hence, despite its elongated structure, the only unique aspect of snR30 compared to modification H/ACA snoRNAs is that its 5′ hairpin does not have any known RNA targets.
Human U17 RNA (207 nt) is shorter than yeast snR30 and comprises four hairpins (Figure 3B), forming a secondary structure consisting of a 5′-variable domain and a 3′-conserved domain [25][74][76]. Thus, the comparison of yeast snR30 and human U17 can reveal functionally important regions of this conserved H/ACA snoRNA. During evolution, the 5′ region of snR30/U17 was compacted to a smaller size, effectively reducing transcriptional cost, which is similar to the general trend of guide RNA shortening between single-cell and complex eukaryotes [77]. In humans, the 5′ end of U17 is composed of two stems of similar size prior to the H box. Whereas the H box of U17 is not required for in vitro RNP formation, the H box of snR30 is critical for accumulation of snR30 in vivo [69][74]. U17 also contains an internal hairpin, although it is much smaller than that of snR30. Since the 5′ structure of snR30/U17 is not conserved, mutational studies investigated whether the 5′ and internal hairpins of yeast snR30 are critical for cell viability [74]. Indeed, both the 5′ hairpin and the internal hairpin can be individually deleted without affecting cell viability, and cell viability was maintained at a reduced level when replacing both the 5′ and internal hairpins with the 5′ hairpin of another box H/ACA RNA. Therefore, the 5′ hairpin and the internal hairpin of yeast snR30 are not directly responsible for its essential role within the cell. In contrast to the 5′ domain, the 3′ hairpin of U17 is extremely similar to that of snR30 as they both possess a structure identical to a standard H/ACA guide RNA hairpin including an unpaired bulge. As further outlined below, the conserved nature of the 3′ hairpin already indicates that this region in snR30/U17 is functionally most important.
By aligning the sequences of the snR30/U17 species from yeast, Xenopus, and humans, two strongly conserved sequence motifs in the 3′ hairpin were discovered and called m1 and m2, which are critical for ribosome formation [74]. The m1 and m2 regions are located in the non-productive, unpaired ‘pseudouridylation pocket’ on the 3′ hairpin of snR30/U17. However, rather than being located on the distal side of the pocket where modification H/ACA guide RNAs bind their targets, they are located on the basal side of the pocket. Two complementary sequences in 18S rRNA were discovered and designated as rm1 and rm2, and mutational studies confirmed Watson-crick base pairing between the m1/rm1 and m2/rm2 sequences that is necessary for pre-rRNA processing [74]. While there is some variation in the m1 and m2 sequences across eukaryotes, these are always matched by compensatory mutations in the rm1 and rm2 motifs in 18S rRNA (Table 1), underlining the importance of this base-pairing of snR30/U17 with 18S rRNA for ribosome biogenesis.
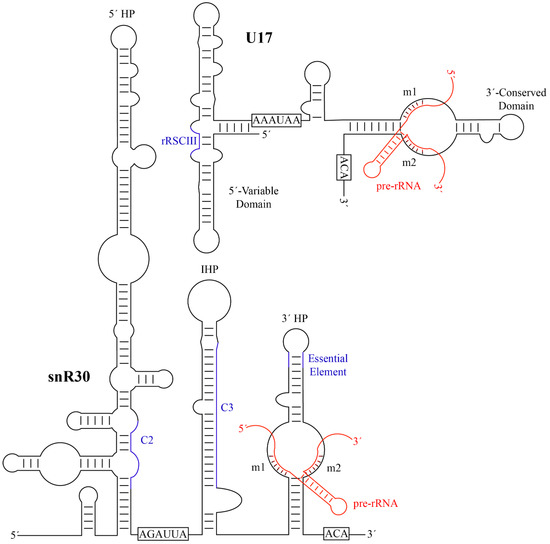
Figure 2. Secondary structures of yeast snR30 and human U17 and their modus operandi of pre-RNA binding. The H and ACA sequences, that characterize each H/ACA snoRNA, are boxed. The m1 and m2 motifs are labelled and the base-pairing to target pre-rRNA is shown in red. Additional regions of snR30/U17 predicted to have a function such as forming further interactions with 18S rRNA are depicted in blue (C2 and C3 in S. cerevisiae snR30, rRSCIII in human U17). HP, hairpin; IHP, internal hairpin. Minor bulges and imperfect base-pairing are not represented. snR30 was adapted from Atzorn et al. [74], and U17 was adapted from Ruhl et al. [78].
In addition to the critical and highly conserved m1 and m2 regions, additional elements of snR30 or U17 have been identified that also bind to 18S rRNA, although these secondary interactions are not conserved across all species. In S. cerevisiae, crosslinking, ligation, and sequencing of hybrids (CLASH) uncovered additional areas of interaction between snR30 and 18S rRNA [34]. Notably, two of the strongest interaction sites, called C2 and C3, also contain regions of sequence complementarity between snR30 and 18S rRNA and are conserved among fungi (Figure 3). First, a region in the 5′ hairpin of snR30 is proposed to interact with helix 1 in expansion segment 6 (ES6) of 18S rRNA in close vicinity to the interaction of the m1 and m2 regions with helix 3 of ES6. However, the importance of this interaction remains to be investigated since the 5′ hairpin of snR30 is dispensable. Second, a 19-nt region within the internal hairpin of snR30 has the potential to base-pair to expansion segment 7 (ES7) in 18S rRNA, which is also conserved in Schizosaccharomyces pombe. Whereas these interactions sites are likely specific to fungi, other putative contacts between U17 and 18S rRNA have been reported in vertebrates. In humans, the U17 rRCSIII sequence in stem 2 of the 5′ domain is predicted to base-pair with 18S rRNA at positions 967–976 (Figure 2) [79], and this sequence complementarity is conserved not only in mammals, but also birds, fish, amphibians, and reptiles. An additional element in stem 1 of the 5′ domain, called rRCSI, may form 12 base-pairs to the 18S rRNA, but is only conserved in fish and amphibians, not in mammals [79]. It remains unknown whether these predicted contacts between vertebrate U17 and 18S rRNA form in vivo and whether they are of functional importance for ribosome biogenesis. Due to the divergence of snR30/U17 sequence and structure over evolution, different interactions with 18S rRNA may have formed that may serve similar functions in stabilizing binding of this H/ACA snoRNA to the SSU processome.
Table 1. Comparison of snR30/U17 RNA across eukaryotes.
| Organism | Common Name | Gene Name | Length (nt) | m1 | m2 | Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| Apteryx rowi | Kiwi | E1/U17 | 205 | AUAUUCCUA | AAACCAU | XR_003255265.1 | N/A |
| Caenorhabditis elegans | Round worm | CeN96 | 221 | AUAUUCCUU | AAACCAU | AY948718.1 | [80] |
| Caretta caretta | Turtle | U17 | 213 | AUAUUCCUA | AAACCAU | AJ306558 | [79] |
| Candida glabrata | Yeast | snR30 | 576 | AUAUUCCUG | AAACCAU | URS00006E7B8C | [81] a |
| Danio rerio | Zebra fish | E1/U17 | >140 | AUAUUCCUA | AAACCAU | LR812563.1 | N/A |
| Fugu rubripes | Pufferfish | U17 | 218 | AUAUUCCUA | AAACCAU | X94942 | [82] |
| Homo sapiens | Humans | U17 | 207 | AUAUUCCUA | AAACCAU | L16791 | [74] |
| Kazachstania naganishii | Yeast | snR30 | 589 | AUAUUCCUA | AAACCAU | URS0000BE6F45 | [81] a |
| Mus musculus | Mouse | U17/SNORA73 | 203 | AUAUUCCUA | AAACCAU | XR_003836540.1 | N/A |
| Pelomedusa subrufa | Turtle | U17 | 215 | AUAUUCCUA | AAACCAU c | AJ306565 | [79] |
| Saccharomyces cerevisiae | Yeast | snR30 | 606 | AUAUUCCUA | AAACCAU | NR_132204 | [74] |
| Saccharomyces pombe | Yeast | U17 | 325 | AUAUUCCUA | AAACCAU | AJ544685 | [74] |
| Tetrahymena thermophila | Algae | U17 | ~240 b | AUAUUCCUG | AAACCAU | AJ544686 | [74] |
| Xenopus laevis | Frog | U17 | 222 | AUAUUCCUA | AAACCAU | X71081 | [54] |
Non-conserved nucleotides in the sequences from C. glabrata and T. thermophila are highlighted in bold. Accession numbers are from GenBank except for S cerevisiae, K. naganishii, and C. glabrata, which are from NCBI or RNA Central. References refer to the publication in which each motif was identified; no reference is provided, indicated as N/A, when the RNA sequence was only identified through a BLAST search at NCBI. a The motif was found manually in the sequences retrieved from RNA Central. b Accession number provides only partial sequence. Atzorn et al. [74] purified and sequenced the 5′ end of T. thermophila snR30, and its length was estimated based on gel size. c The Psu sequence has two A insertions 5′ of the motif, however, the rRNA motif stays the same, indicating that the insertion does not affect base pairing.
References
- Eliceiri, G.L. The vertebrate E1/U17 small nucleolar ribonucleoprotein particle. J. Cell. Biochem. 2006, 98, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Kister, K.P.; Muller, B.; Eckert, W.A. Complex endonucleolytic cleavage pattern during early events in the processing of pre-rRNA in the lower eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1983, 11, 3487–3502. [Google Scholar] [CrossRef] [PubMed]
- Hadjiolova, K.V.; Nicoloso, M.; Mazan, S.; Hadjiolov, A.A.; Bachellerie, J.P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993, 212, 211–215. [Google Scholar] [CrossRef]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Phipps, K.R.; Charette, J.; Baserga, S.J. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev. RNA 2011, 2, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kiss-Laszlo, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef]
- Ni, J.; Tien, A.L.; Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997, 89, 565–573. [Google Scholar] [CrossRef]
- Tollervey, D.; Kiss, T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997, 9, 337–342. [Google Scholar] [CrossRef]
- Lafontaine, D.L.; Tollervey, D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 1999, 5, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.; Tollervey, D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell Biol. 2000, 20, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Segault, V.; Charpentier, B.; Nottrott, S.; Fabrizio, P.; Bachi, A.; Wilm, M.; Rosbash, M.; Branlant, C.; Luhrmann, R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 2000, 103, 457–466. [Google Scholar] [CrossRef]
- Girard, J.P.; Lehtonen, H.; Caizerguesferrer, M.; Amalric, F.; Tollervey, D.; Lapeyre, B. Gar1 is an essential small nucleolar rnp protein required for pre-ribosomal-rna processing in yeast. EMBO J. 1992, 11, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J.; Bousquet-Antonelli, C.; Henry, Y.; Caizergues-Ferrer, M.; Tollervey, D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Gene Dev. 1998, 12, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Henras, A.; Henry, Y.; Bousquet-Antonelli, C.; Noaillac-Depeyre, J.; Gelugne, J.P.; Caizergues-Ferrer, M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998, 17, 7078–7090. [Google Scholar] [CrossRef]
- Zebarjadian, Y.; King, T.; Fournier, M.J.; Clarke, L.; Carbon, J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 1999, 19, 7461–7472. [Google Scholar] [CrossRef]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef]
- Baudin-Baillieu, A.; Fabret, C.; Liang, X.H.; Piekna-Przybylska, D.; Fournier, M.J.; Rousset, J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009, 37, 7665–7677. [Google Scholar] [CrossRef]
- Piekna-Przybylska, D.; Przybylski, P.; Baudin-Baillieu, A.; Rousset, J.P.; Fournier, M.J. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008, 283, 26026–26036. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 2007, 28, 965–977. [Google Scholar] [CrossRef] [PubMed]
- King, T.H.; Liu, B.; McCully, R.R.; Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 2003, 11, 425–435. [Google Scholar] [CrossRef]
- Tollervey, D.; Guthrie, C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985, 4, 3873–3878. [Google Scholar] [CrossRef] [PubMed]
- Bally, M.; Hughes, J.; Cesareni, G. Snr30—A new, essential small nuclear-rna from saccharomyces-cerevisiae. Nucleic Acids Res. 1988, 16, 5291–5303. [Google Scholar] [CrossRef]
- Morrissey, J.P.; Tollervey, D. Yeast Snr30 is a small nucleolar rna required for 18s ribosomal-rna synthesis. Mol. Cell. Biol. 1993, 13, 2469–2477. [Google Scholar] [CrossRef]
- Jinn, S.; Brandis, K.A.; Ren, A.; Chacko, A.; Dudley-Rucker, N.; Gale, S.E.; Sidhu, R.; Fujiwara, H.; Jiang, H.; Olsen, B.N.; et al. snoRNA U17 regulates cellular cholesterol trafficking. Cell Metab. 2015, 21, 855–867. [Google Scholar] [CrossRef]
- Dutca, L.M.; Gallagher, J.E.; Baserga, S.J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011, 39, 5164–5180. [Google Scholar] [CrossRef]
- Brink, M.F.; Verbeet, M.P.; de Boer, H.A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993, 12, 3987–3996. [Google Scholar] [CrossRef]
- Hughes, J.M.; Ares, M., Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5’ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991, 10, 4231–4239. [Google Scholar] [CrossRef]
- Li, H.D.; Zagorski, J.; Fournier, M.J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1990, 10, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Fournier, M.J. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes Dev. 1995, 9, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.P.; Tollervey, D. U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma 1997, 105, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Hackert, P.; Ruprecht, M.; Simm, S.; Bruning, L.; Mirus, O.; Sloan, K.E.; Kudla, G.; Schleiff, E.; Bohnsack, M.T. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 2014, 20, 1173–1182. [Google Scholar] [CrossRef]
- Tollervey, D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987, 6, 4169–4175. [Google Scholar] [CrossRef]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017, 24, 954–964. [Google Scholar] [CrossRef]
- Barandun, J.; Chaker-Margot, M.; Hunziker, M.; Molloy, K.R.; Chait, B.T.; Klinge, S. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017, 24, 944–953. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Barandun, J.; Hunziker, M.; Klinge, S. Architecture of the yeast small subunit processome. Science 2017, 355. [Google Scholar] [CrossRef]
- Dragon, F.; Gallagher, J.E.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef]
- Grandi, P.; Rybin, V.; Bassler, J.; Petfalski, E.; Strauss, D.; Marzioch, M.; Schafer, T.; Kuster, B.; Tschochner, H.; Tollervey, D.; et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 2002, 10, 105–115.
- Zhang, L.M.; Wu, C.; Cai, G.H.; Chen, S.; Ye, K.Q. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Gene Dev. 2016, 30, 718–732. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Hunziker, M.; Barandun, J.; Dill, B.D.; Klinge, S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015, 22, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Chaker-Margot, M.; Klinge, S. Assembly and early maturation of large subunit precursors. RNA 2019, 25, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xie, Z.; Yang, F.; Ye, K. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 2017, 45, 6837–6847. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fernandez, J.; Roman, A.; De Las Rivas, J.; Bustelo, X.R.; Dosil, M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007, 27, 5414–5429. [Google Scholar] [CrossRef] [PubMed]
- Kornprobst, M.; Turk, M.; Kellner, N.; Cheng, J.; Flemming, D.; Kos-Braun, I.; Kos, M.; Thoms, M.; Berninghausen, O.; Beckmann, R.; et al. Architecture of the 90S pre-ribosome: A structural view on the birth of the eukaryotic ribosome. Cell 2016, 166, 380–393. [Google Scholar] [CrossRef]
- Cheng, J.; Bassler, J.; Fischer, P.; Lau, B.; Kellner, N.; Kunze, R.; Griesel, S.; Kallas, M.; Berninghausen, O.; Strauss, D.; et al. Thermophile 90S pre-ribosome structures reveal the reverse order of co-transcriptional 18S rRNA subdomain integration. Mol. Cell 2019, 75, 1256–1269.e1257. [Google Scholar] [CrossRef]
- Ursic, D.; Himmel, K.L.; Gurley, K.A.; Webb, F.; Culbertson, M.R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997, 25, 4778–4785. [Google Scholar] [CrossRef]
- Steinmetz, E.J.; Brow, D.A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell Biol. 1996, 16, 6993–7003. [Google Scholar] [CrossRef]
- Chanfreau, G.; Legrain, P.; Jacquier, A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 1998, 284, 975–988.
- Mouaikel, J.; Verheggen, C.; Bertrand, E.; Tazi, J.; Bordonne, R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 2002, 9, 891–901. [Google Scholar] [CrossRef]
- Kiss, T.; Filipowicz, W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993, 12, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Pelczar, P.; Filipowicz, W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5’-terminal oligopyrimidine gene family. Mol. Cell Biol. 1998, 18, 4509–4518. [Google Scholar] [CrossRef]
- Cecconi, F.; Mariottini, P.; Loreni, F.; Pierandrei-Amaldi, P.; Campioni, N.; Amaldi, F. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a sequence repeated in the six introns of Xenopus laevis ribosomal protein S8 gene. Nucleic Acids Res. 1994, 22, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Filipowicz, W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995, 9, 1411–1424. [Google Scholar] [CrossRef]
- Cecconi, F.; Mariottini, P.; Amaldi, F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995, 23, 4670–4676. [Google Scholar] [CrossRef]
- Szczepinska, T.; Kalisiak, K.; Tomecki, R.; Labno, A.; Borowski, L.S.; Kulinski, T.M.; Adamska, D.; Kosinska, J.; Dziembowski, A. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015, 25, 1622–1633. [Google Scholar] [CrossRef]
- Caffarelli, E.; Fatica, A.; Prislei, S.; De Gregorio, E.; Fragapane, P.; Bozzoni, I. Processing of the intron-encoded U16 and U18 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996, 15, 1121–1131. [Google Scholar] [CrossRef]
- Kufel, J.; Grzechnik, P. Small nucleolar RNAs Tell a different tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef]
- Yang, P.K.; Rotondo, G.; Porras, T.; Legrain, P.; Chanfreau, G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002, 277, 45235–45242.
- Walbott, H.; Machado-Pinilla, R.; Liger, D.; Blaud, M.; Rety, S.; Grozdanov, P.N.; Godin, K.; van Tilbeurgh, H.; Varani, G.; Meier, U.T.; et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011, 25, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Caton, E.A.; Kelly, E.K.; Kamalampeta, R.; Kothe, U. Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 2018, 46, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Machado-Pinilla, R.; Liger, D.; Leulliot, N.; Meier, U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 2012, 18, 1833–1845. [Google Scholar] [CrossRef]
- Darzacq, X.; Kittur, N.; Roy, S.; Shav-Tal, Y.; Singer, R.H.; Meier, U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006, 173, 207–218. [Google Scholar] [CrossRef]
- Grozdanov, P.N.; Roy, S.; Kittur, N.; Meier, U.T. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA 2009, 15, 1188–1197. [Google Scholar] [CrossRef]
- Narayanan, A.; Lukowiak, A.; Jady, B.E.; Dragon, F.; Kiss, T.; Terns, R.M.; Terns, M.P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999, 18, 5120–5130. [Google Scholar] [CrossRef]
- Lange, T.S.; Ezrokhi, M.; Amaldi, F.; Gerbi, S.A. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 1999, 10, 3877–3890. [Google Scholar] [CrossRef]
- Dragon, F.; Pogacic, V.; Filipowicz, W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell Biol. 2000, 20, 3037–3048. [Google Scholar] [CrossRef]
- Samarsky, D.A.; Fournier, M.J.; Singer, R.H.; Bertrand, E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998, 17, 3747–3757. [Google Scholar] [CrossRef]
- Narayanan, A.; Speckmann, W.; Terns, R.; Terns, M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 1999, 10, 2131–2147. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Fayet, E.; Jady, B.E.; Richard, P.; Weber, M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Meier, U.T. RNA modification in Cajal bodies. RNA Biol. 2017, 14, 693–700.
- Atzorn, V.; Fragapane, P.; Kiss, T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell Biol. 2004, 24, 1769–1778. [Google Scholar] [CrossRef]
- Ganot, P.; Bortolin, M.L.; Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef]
- Cervelli, M.; Oliverio, M.; Bellini, A.; Bologna, M.; Cecconi, F.; Mariottini, P. Structural and sequence evolution of U17 small nucleolar RNA (snoRNA) and its phylogenetic congruence in chelonians. J. Mol. Evol. 2003, 57, 73–84. [Google Scholar] [CrossRef]
- Hughes, J.M.; Konings, D.A.; Cesareni, G. The yeast homologue of U3 snRNA. EMBO J. 1987, 6, 2145–2155. [Google Scholar] [CrossRef]
- Ruhl, D.D.; Pusateri, M.E.; Eliceiri, G.L. Multiple conserved segments of E1 small nucleolar RNA are involved in the formation of a ribonucleoprotein particle in frog oocytes. Biochem. J. 2000, 348, 517–524. [Google Scholar] [CrossRef]
- Cervelli, M.; Cecconi, F.; Giorgi, M.; Annesi, F.; Oliverio, M.; Mariottini, P. Comparative structure analysis of vertebrate U17 small nucleolar RNA (snoRNA). J. Mol. Evol. 2002, 54, 166–179. [Google Scholar] [CrossRef]
- Deng, W.; Zhu, X.; Skogerbo, G.; Zhao, Y.; Fu, Z.; Wang, Y.; He, H.; Cai, L.; Sun, H.; Liu, C.; et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: Genomic features, biogenesis, and expression. Genome Res. 2006, 16, 20–29. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, F.; Crosio, C.; Mariottini, P.; Cesareni, G.; Giorgi, M.; Brenner, S.; Amaldi, F. A functional role for some Fugu introns larger than the typical short ones: The example of the gene coding for ribosomal protein S7 and snoRNA U17. Nucleic Acids Res. 1996, 24, 3167–3172.




