
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Huali Zhang | + 1622 word(s) | 1622 | 2021-12-01 08:45:43 | | | |
| 2 | Catherine Yang | Meta information modification | 1622 | 2021-12-10 09:38:08 | | |
Video Upload Options
Starch is the main food source for human beings and livestock all over the world, and it is also the raw material for production of industrial alcohol and biofuel. Because of the complexity and flexibility of carbon allocation in the formation of endosperm starch, cereal crops require a broad range of enzymes and one matching network of regulators to control the providential functioning of these starch biosynthetic enzymes.
1. Introduction
As a fundamental commodity, starch was and is still widely used for human consumption in food and non-food industries [1][2]. Since starch is the major storage carbohydrate and generally accumulated in the heterotrophic starch-storing organs of crops [3][4][5], a large amount of starch is sourced from the attainable parts of staple crop plants [5], especially for cereal seeds, roots, and tubers. Among them, cereal seeds make up most of the worldwide annual starch production.
Starch is composed of amylose and amylopectin glucan polymers, which are packaged to form the insoluble semi-crystalline starch granules [6]. Starch synthesized in cereal crops contains at least two types, termed transitory starch and storage starch. Transitory starch is usually found in the plastid of photosynthetic organs and displays circadian turnover regulation with diurnal cycles [7]. While the storage starch is synthesized in the amyloplasts of non-photosynthetic sink tissues (i.e., seed endosperm), and requires the supply of sucrose and ATP from the source organs (i.e., leaves) [5]. Interestingly, some common regulatory factors of starch synthesis exist between chloroplasts and amyloplasts. For example, the expression of starch biosynthetic genes, i.e., GBSSI , was partly mediated by tetrapyrrole intermediates, i.e., heme, in endosperm during early seed development in rice [8], suggesting potential shared regulatory networks in various plastids.
The process of starch biosynthesis in crops requires tight cooperation of various starch biosynthetic enzymes, and coordinates with other metabolisms. Until now, a great number of genes involved in starch biosynthesis have been identified in various cereals. However, despite these findings, the regulatory factors underlying starch synthesis are still poorly understood, especially regarding the regulation of starch biosynthetic genes. Therefore, the screening and identification of key regulators is vital to understanding the fundamental regulatory mechanism underlying starch synthesis.
2. Transcription Factors Directly Regulating Starch Biosynthesis
Interestingly, ZmbZIP91 [9] and O2 [10] are the two closest homologous bZIP transcription factors of OsbZIP58. However, their function seems not to be fully in line with OsZIP58, and has divergent target genes and tissue dependence. ZmbZIP91 could bind to the ACTCAT directly to regulate the expression of AGPS1 , SSI , SSIIIa , and ISA1 during starch synthesis [9], while O2 [10] usually combines with other factors in the regulation of starch biosynthesis ( Figure 1 ).
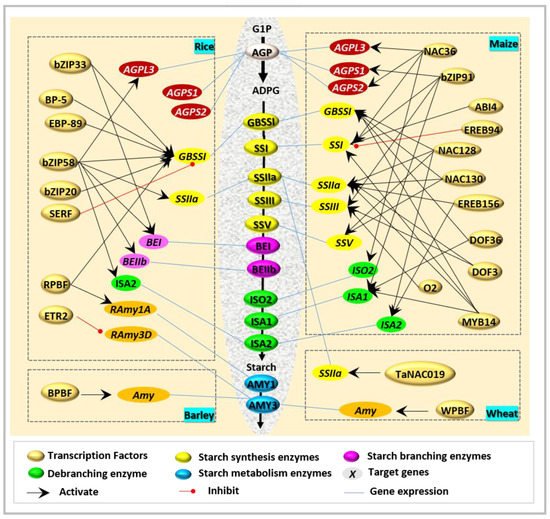
Recently, a novel NAC-type transcription factor was revealed to regulate starch biosynthesis in wheat. TaNAC019 is specifically expressed in endosperm, and functions during starch synthesis through the following ways [11]: (i) by direct binding to the protomers of glutenin-encoded genes, (ii) by mediation of the accumulation of storage proteins via regulated Wheat storage protein activator ( TaSPA ) expression and interaction with TaGAMyb, and (iii) by adjustment of the accumulation of starch by the regulation of SSIIa and Susy1.
As the widely distributed transcription factors, MYBs are vital for a variety of biological functions. However, the functional studies of MYBs on starch biosynthesis are still scarce in cereal crops. So far, only one MYB factor has been reported as a regulator during starch biosynthesis [12].
GRAS transcription factors are involved in plant growth and development processes, especially for endodermis specification [13]. Besides, GRAS is greatly associated with type I genes during endosperm starch synthesis in rice [14], but its function as a regulator for starch biosynthesis is still not understood in cereal crops.
3. Other Regulators Directly Affecting Starch Biosynthesis
FLOURY ENDOSPREM is named based on its well-recognized but viable endosperm phenotypes, which usually display a chalky and soft endosperm. So far, 16 rice floury mutants [15] and three maize floury mutants [16] have been identified. However, only rice FLO2 [17] and FLO7 [18], as well as ZmFloury3 [16] serve as regulators of starch biosynthesis but do not act as enzymes. Although all of them play key roles in the regulation of starch formation, and are also responsible for aberrant seeds with reduced grain quality [16][17][18], their functioning models vary.
FLO2 acts as a regulatory protein that controls the biosynthesis of seed storage substances, possibly through its tetratricopeptide repeat (TPR) motifs for protein–protein interactions [17]. At least two proteins have been reported that interact with FLO2 during starch synthesis ( Figure 2 ). One basic helix–loop–helix (bHLH) protein may interact with FLO2 to modulate the expression of starch biosynthesis-associated nuclear genes (SBANGs) [17]. Subsequently, the FLO2-interacting cupin domain protein 1 (FLOC1) was reported to interact with the TPR motif of FLO2 to maintain fertility and seed quality in rice [19], even though they are supposed to have different functions.
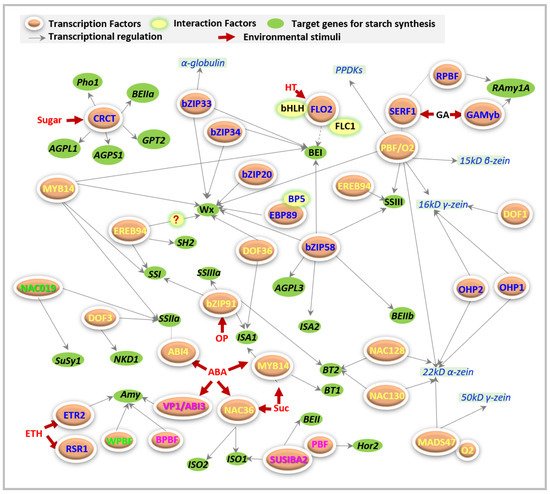
FLO7 prefers to specifically function in the endosperm periphery, and harbors two essential domains of an N -terminal transit peptide and an unknown function 1338 (DUF1338), during starch synthesis and amyloplast development [18].
The CRCT protein has been reported to be a regulator of starch biosynthesis in rice but does not seem to belong to a zinc finger motif [20], and cannot be listed in any of the above groups. Thus, here, CRCT is grouped independently. The expression of OsCRCT displays diurnal oscillation and is mainly focused on the vegetative organs. Therefore, OsCRCT serves as a positive regulator of several starch biosynthetic genes, including AGPS1 , AGPL1 , Glc-6-phosphate translocator2 ( GPT2 ) , plastidial α-glucan phosphorylase1 ( Pho1 ), and BEIIa , and contributes to starch biosynthesis in rice leaves [20]. The effects of OsCRCT on the storage of starch are unclear so far. However, the use of plants overexpressing OsCRCT is a potential approach to improve yields for food and biofuel [20], depending on the increased contents of starch derived from enhanced synthesis of starch instead of the defects of starch breakdown or translocation.
4. Regulators Indirectly Mediating Starch Biosynthesis
Aside from the direct regulators of starch biosynthesis, many other factors also participate in the regulation of starch synthesis, partially through the post-transcriptional regulation of TFs, i.e., micro RNAs (miRNAs), or via regulators of the formation of the upstream precursors and/or downstream products of starch.
Some transcription factors have been reported to mediate starch accumulation in cereal endosperms, but they participate in regulation via control of the synthesis of the downstream and/or upstream products of starch, i.e., sugar and proteins, instead of directly regulating starch synthesis. Here, we mainly concentrate on some of them ( Table 1 ), including bZIPs, NAC, DOF, MADS, and basic helix-loop-helix (bHLH).
Recently, one B3 domain-containing AP2/ERF transcriptional factor, ZmABI19 [21], was well documented to mediate starch synthesis via binding with RY through the regulation of cooperated expression of O2 , PBF1 , ZmbZIP22 , NAC130 , and O11 .
Regulation of NAC on starch synthetic enzymes is often accompanied with a simultaneous regulation of storage protein synthesis-associated genes ( Table 1 ). Although ZmNAC128 and ZmNAC130 have been well documented to regulate several starch biosynthetic genes, they can also bind with the motif of ACGCAA to activate the expression of 16-kDa γ-zein genes [22]. However, these two NAC factors [22] do not dimerize with each other but are functionally redundant for starch and protein accumulation. A similar phenomenon was also found in wheat [11]. TaNAC019 [11] has been reported to directly bind to the promoters of glutenin-encoded genes and be involved in the accumulation of starch and proteins. Therefore, the regulation of starch biosynthesis is not independent but prefers to act synergistically with other metabolic processes, i.e., protein synthesis.
Table 1. Regulators indirectly affecting starch biosynthesis of cereal crops.
| TF | Binding Domains | Target Genes | Specific Expressed Tissues | References |
|---|---|---|---|---|
| Oryza sativa L. | ||||
| RISBZ1/bZIP58 | TCCACGT(a/c)R(a/t) and GATGYRTGG | O2 | Endosperm | [23] |
| Zea mays L. | ||||
| ZmbZIP22 | ACAGCTCA | 27-kDa γ-zein | Endosperm | [24] |
| O2 | GA/TGAPyPuTGPu | PPDK | [25][26][27] | |
| O2 | TCCACGTAGA | 22 kDa zein | All types of tissues | [25][26][27] |
| OHP1 | O2-likebox (TTTACGT) | 27-kDa γ-zein and 22-kDa α-zein | [28] | |
| OHP2 | O2-likebox (TTTACGT) | zein | [28] | |
| ZmABI19 | RY | O2, PBF1, ZmbZIP22, NAC130, O11 | Endosperm and embryo | [21] |
| ZmNAC128 | ACGCAA | 16-kDa γ-zein | Endosperm | [22] |
| ZmNAC130 | ACGCAA | 16-kDa γ-zein | Endosperm | [22] |
| ZmMADS47 | CATGT | α-zein and 50-kDa γ-zein | Endosperm | [29] |
| PBF/DOF13 | TGTAAAG | 27-kD γ- and 22-kD α-zein | Endosperm | [10] |
| DOF1 | γ-zein | Endosperm | [30] | |
| Triticum aestivum L. | ||||
| TaNAC019 | SPA, GaMyb, Glutenin | Endosperm | [11] | |
References
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011, 155, 1566–1577.
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010, 61, 209–234.
- Sulpice, R.; Pyl, E.T.; Ishihara, H.; Trenkamp, S.; Steinfath, M.; Witucka-Wall, H.; Gibon, Y.; Usadel, B.; Poree, F.; Piques, M.C. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 2009, 106, 10348–10353.
- Geigenberger, P.; Stitt, M.; Fernie, A.R. Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ. 2010, 27, 655–673.
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106.
- Sabelli, P.A.; Larkins, B.A. The development of endosperm in grasses. Plant Physiol. 2009, 149, 14–26.
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292.
- Li, R.; Jiang, M.; Zheng, W.; Zhang, H. GUN4-mediated tetrapyrrole metabolites regulates starch biosynthesis during early seed development in rice. J. Cereal Sci. 2021, 101, 103317.
- Chen, J.; Yi, Q.; Cao, Y.; Wei, B.; Zheng, L.; Xiao, Q.; Xie, Y.; Gu, Y.; Li, Y.; Huang, H.; et al. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 2016, 67, 1327–1338.
- Zhang, Z.; Zheng, X.; Yang, J.; Messing, J.; Wu, Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 10842–10847.
- Gao, Y.; An, K.; Guo, W.; Chen, Y.; Zhang, R.; Zhang, X.; Chang, S.; Rossi, V.; Jin, F.; Cao, X.; et al. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat gain quality. Plant Cell 2021, 33, 603–622.
- Xiao, Q.; Wang, Y.; Du, J.; Li, H.; Wei, B.; Wang, Y.; Li, Y.; Yu, G.; Liu, H.; Zhang, J.; et al. ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT1 promoter in starch biosynthesis in maize. FEBS J. 2017, 284, 3079–3099.
- Wang, X.L.; Chen, Y.P.; De-yue, Y. Expression of the MADS-Box gene GmAGL15 in seed development of soybean: Expression the MADS-Box gene GmAGL15 in seed development of Soybean. Acta Agron. Sin. 2008, 34, 330–332.
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938.
- Teng, X.; Zhong, M.; Zhu, X.; Wang, C.; Ren, Y.; Wang, Y.; Zhang, H.; Jiang, L.; Wang, D.; Hao, Y.; et al. FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice. Plant Biotechnol. J. 2019, 17, 1914–1927.
- Li, Q.; Wang, J.; Ye, J.; Zheng, X.; Xiang, X.; Li, C.; Fu, M.; Wang, Q.; Zhang, Z.; Wu, Y. The maize imprinted gene Floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III. Plant Cell 2017, 29, 2661–2675.
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294.
- Zhang, L.; Ren, Y.; Lu, B.; Yang, C.; Feng, Z.; Liu, Z.; Chen, J.; Ma, W.; Wang, Y.; Yu, X.; et al. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J. Exp. Bot. 2016, 67, 633–647.
- Suzuki, R.; Imamura, T.; Nonaga, Y.; Kusano, H.; Teramura, H.; Sekine, K.T.; Yamashita, T.; Shimada, H. A novel FLOURY ENDOSPERM2 (FLO2)-interacting protein, is involved in maintaining fertility and seed quality in rice. Plant Biotechnol. J. 2020, 37, 47–55.
- Morita, R.; Sugino, M.; Hatanaka, T.; Misoo, S.; Fukayama, H. CO2-responsive CONSTANS, CONSTANS-like, and time of chlorophyll a/b binding protein Expression1 protein is a positive regulator of starch synthesis in vegetative organs of rice. Plant Physiol. 2015, 167, 1321–1331.
- Yang, T.; Guo, L.; Ji, C.; Wang, H.; Wang, J.; Zheng, X.; Xiao, Q.; Wu, Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 2020, koaa008.
- Zhang, Z.; Dong, J.; Chen, J.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228.
- Onodera, Y.; Suzuki, A.; Wu, C.; Washida, H.; Takaiwa, F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J. Biol. Chem. 2001, 276, 14139–14152.
- Li, C.; Yue, Y.; Chen, H.; Qi, W.; Song, R. The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 2018, 30, 2402–2424.
- Stower, H. Gene regulation: Resolving transcription factor binding. Nat. Rev. Genet. 2011, 13, 71.
- Lohmer, S.; Maddaloni, M.; Motto, M.; Di Fonzo, N.; Hartings, H.; Salamini, F.; Thompson, R.D. The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J. 1991, 10, 617.
- Schmidt, R.J.; Ketudat, M.; Aukerman, M.J.; Hoschek, G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 1992, 4, 689–700.
- Yang, J.; Ji, C.; Wu, Y. Divergent Transactivation of Maize Storage Protein Zein Genes by the Transcription Factors Opaque2 and OHPs. Genetics 2016, 204, 581–591.
- Dong, Q.; Wang, F.; Kong, J.; Xu, Q.; Li, T.; Chen, L.; Chen, H.; Jiang, H.; Li, C.; Cheng, B. Functional analysis of ZmMADS1a reveals its role in regulating starch biosynthesis in maize endosperm. Sci. Rep. 2019, 9, 3253.
- Washio, K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein–protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003, 133, 850–863.




