
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bruno Casciaro | + 876 word(s) | 876 | 2020-06-18 11:19:33 | | | |
| 2 | Nicole Yin | -7 word(s) | 869 | 2020-11-05 10:43:12 | | |
Video Upload Options
The discovery of antibiotics has revolutionized the medicine and treatment of microbial infections. However, the current scenario has highlighted the difficulties in marketing new antibiotics and an exponential increase in the appearance of resistant strains. Here, the main antibiotic resistance mechanisms are briefly listed with some examples.
1. Introduction
Biologically active molecules are defined as antimicrobials if they are able to inhibit growth or to kill certain or various classes of microorganisms. However, microorganisms have developed several mechanisms to circumvent the action of these antimicrobial compounds. In this context, it is worthwhile to specify the difference between antimicrobial resistance and persistence. Resistance to a given molecule is maintained from the mother cell to the daughter cells, unless mutations make them susceptible again [1]. In comparison, persistence is defined as the ability of microbial cells to be recalcitrant to the antibiotic action, as they enter into a stationary phase of their growth (dormant cells). This leads to the inefficacy of the antibiotic agents since most of them act by inhibiting or interacting with specific metabolic processes that are not active in dormant cells [2][3]. It is also important to define the two major types of antimicrobial resistance: natural and acquired [4][5]. Natural resistance can be constantly expressed in the bacterial species (intrinsic resistance), whereas acquired resistance is expressed only upon exposure to an antimicrobial agent (induced resistance) [6]. The reduced permeability of the outer membrane and the activity of efflux pumps are classic examples of intrinsic resistance [7]. Acquired resistance occurs through acquisition of genetic material by means of transformation, conjugation, transposition (horizontal gene transfer) or by mutation in the chromosomal DNA [8][9]. Mutation of the drug target or in those genes involved in the regulation of drug transporters are examples of acquired resistance [4].
2. Mechanisms of Antimicrobial Resistance
Gram(+) and Gram(−) bacteria possess and/or have developed several mechanisms of antimicrobial resistance that fall into five major categories (Figure 1):
Limitation of drug uptake. Bacteria can be intrinsically resistant to a certain antimicrobial due to their structure and morphology. Lipopolysaccharide (LPS) in Gram(−) bacteria, for example, provides a physical barrier that protects the cell from several groups of large molecules [10]. In these bacteria, drugs are internalized through porin channels that generally allow the uptake of hydrophilic molecules. Mutations that change their selectivity or that reduce the number of expressed porins are the two major mechanisms of antimicrobial resistance [11][12]. Gram(+) bacteria, lacking outer membrane, possess a peptidoglycan cell wall and the restricting drug uptake is not as prevalent. However, pathogenic bacterial species, i.e., Staphylococcus aureus, have developed a mechanism which consists in thickening the cell wall to limit the amount of drug that enters the cell [13][14]. Mycoplasma spp, devoid of cell wall, are intrinsically resistant to antimicrobials (e.g., β-lactams and glycopeptides) that interfere with cell wall synthesis and regulation [15].
Drug inactivation. Bacteria can produce several enzymes or molecules that inactivate drugs by covalent binding or enzymatic processes. Firstly, common antibiotics (e.g., aminoglycosides, streptogramins, fluoroquinolones, chloramphenicol) could be inactivated by acetylation, phosphorylation or adenylation; secondly, hydrolyzation is the primary mechanism by which bacteria can inactivate β-lactam antibiotics (e.g., cephalosporins, penicillins and cephamycins). β-lactamases are the most common example: these enzymes provide resistance to β-lactam antibiotics by hydrolyzing a specific site in the β-lactam ring structure [16]. Recently, β-lactamases were found to be active against carbapenems in Enterobacteriaceae (carbapenemases, i.e., Klebsiella pneumoniae carbapenemases and carbapenem-resistant enterobacteriaceae enzymes) [16].
Mutation/alteration of the drug target. The majority of antimicrobials have a specific mechanism of action against a specific cellular target and this is one of the reasons why bacteria are not susceptible to a certain class of molecules [17]. Gram(+) strains, for instance, become resistant to β-lactam drugs via alteration of the penicillin-binding proteins, that are transpeptidases involved in the cell wall construction [18]. S. aureus acquires resistance to the glycopeptide vancomycin by decreasing the binding ability of this molecule to the cell wall, as a consequence of a modification of the terminal d-Ala–d-Ala moiety of the peptidoglycan precursor lipid II [19].
Drug efflux. Bacteria can eliminate internalized toxic substances through a mechanism involving efflux pumps, which can be constitutively expressed or overexpressed under certain conditions. Many of these pumps have the capability to transport different types of substances. They are properly named multi-drug efflux pumps [20] and their increased number is generally associated to high-level of resistance to clinically significant bacterial infections [21][22].
Biofilm formation. In conditions of environmental stress, scarcity of nutrients, presence of antimicrobial molecules, some bacterial species can switch from a motile to a sessile lifestyle, named biofilm. This is a bacterial community able to colonize abiotic (e.g., medical devices and implants [23][24][25]) and biotic surfaces (e.g., human tissues [26][27][28]). Biofilm formation is a strategy used by pathogenic bacteria to protect themselves from the external stressful conditions by producing a thick and sticky extracellular matrix which contains DNA, proteins and polysaccharides. In addition, biofilm cells enter into a slow division rate, which weakens the effect of antibiotic molecules targeting specific cellular processes. Thus, biofilms are often associated to chronic infections and molecules capable to disrupt these communities and/or to inhibit their formation are highly demanded [29][30].
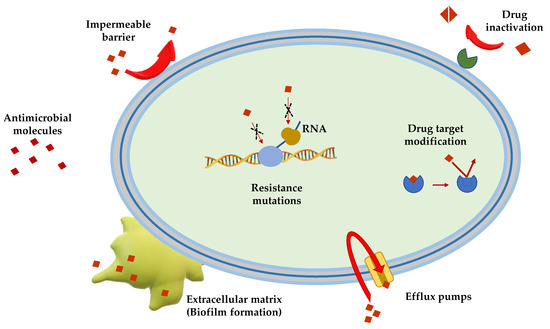
Figure 1. Schematic representation of the principal mechanisms of antibiotic resistance.
References
- Wanda Reygaert; An overview of the antimicrobial resistance mechanisms of bacteria.. AIMS Microbiology 2018, 4, 482-501, 10.3934/microbiol.2018.3.482.
- Thomas K. Wood; Stephen J. Knabel; Brian W. Kwan; Bacterial Persister Cell Formation and Dormancy. Applied and Environmental Microbiology 2013, 79, 7116-7121, 10.1128/AEM.02636-13.
- Iris Keren; Niilo Kaldalu; Amy Spoering; Yipeng Wang; Kim Lewis; Persister cells and tolerance to antimicrobials. FEMS Microbiology Letters 2004, 230, 13-18, 10.1016/s0378-1097(03)00856-5.
- José Luis Martínez; General principles of antibiotic resistance in bacteria. Drug Discovery Today: Technologies 2014, 11, 33-39, 10.1016/j.ddtec.2014.02.001.
- Mohsen Arzanlou; Wern Chern Chai; Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays in Biochemistry 2017, 61, 49-59, 10.1042/ebc20160063.
- Georgina Cox; Gerard D. Wright; Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. International Journal of Medical Microbiology 2013, 303, 287-292, 10.1016/j.ijmm.2013.02.009.
- Jessica M. A. Blair; Mark A Webber; Alison J. Baylay; David O. Ogbolu; Laura J. V. Piddock; Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology 2014, 13, 42-51, 10.1038/nrmicro3380.
- Bogdan Ioan Coculescu; Antimicrobial resistance induced by genetic changes. Journal of Medicine and Life 2009, 2, 114-123.
- Julian Davies; Dorothy Davies; Origins and Evolution of Antibiotic Resistance. Microbiology and Molecular Biology Reviews 2010, 74, 417-433, 10.1128/mmbr.00016-10.
- Rita F. Maldonado; Isabel Sá-Correia; Miguel A. Valvano; Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiology Reviews 2016, 40, 480-493, 10.1093/femsre/fuw007.
- Jessica M. A. Blair; Grace E Richmond; Laura J. V. Piddock; Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiology 2014, 9, 1165-1177, 10.2217/fmb.14.66.
- Umji Choi; Chang-Ro Lee; Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli.. Frontiers in Microbiology 2019, 10, 953, 10.3389/fmicb.2019.00953.
- Longzhu Cui; Xiaoxue Ma; Katsuhiro Sato; Keiko Okuma; Fred C. Tenover; Elsa M. Mamizuka; Curtis G. Gemmell; M.-N. Kim; Marie-Cecile Ploy; N. El Solh; et al.Vivian FerrazKeiichi Hiramatsu Cell Wall Thickening Is a Common Feature of Vancomycin Resistance in Staphylococcus aureus. Journal of Clinical Microbiology 2003, 41, 5-14, 10.1128/JCM.41.1.5-14.2003.
- Ana Belén García; José Manuel Viñuela-Prieto; Laura López-González; Francisco Javier Candel; Correlation between resistance mechanisms in Staphylococcus aureus and cell wall and septum thickening. Infection and Drug Resistance 2017, 10, 353-356, 10.2147/IDR.S146748.
- Cecile Bebear; Sabine Pereyre; Olivia Peuchant; Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiology 2011, 6, 423-431, 10.2217/fmb.11.18.
- Catherine L. Tooke; Philip Hinchliffe; Eilis C. Bragginton; Charlotte K. Colenso; Viivi H. A. Hirvonen; Yuiko Takebayashi; James Spencer; β-Lactamases and β-Lactamase Inhibitors in the 21st Century. Journal of Molecular Biology 2019, 431, 3472-3500, 10.1016/j.jmb.2019.04.002.
- Bruno Casciaro; Maria Rosa Loffredo; Vincenzo Luca; Walter Verrusio; Mauro Cacciafesta; Maria Luisa Mangoni; Esculentin-1a Derived Antipseudomonal Peptides: Limited Induction of Resistance and Synergy with Aztreonam. Protein & Peptide Letters 2018, 25, 1155-1162, 10.2174/0929866525666181101104649.
- Jae-Hee Jeong; Hyung Jin Cha; Yeon-Gil Kim; Crystal Structures of Penicillin-Binding Protein D2 fromListeria monocytogenesand Structural Basis for Antibiotic Specificity. Antimicrobial Agents and Chemotherapy 2018, 62, AAC.00796-18, 10.1128/aac.00796-18.
- Yanguang Cong; Sijin Yang; Xiancai Rao; Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. Journal of Advanced Research 2020, 21, 169-176, 10.1016/j.jare.2019.10.005.
- Sepideh Hassanzadeh; Sara Ganjloo; Mohammad Reza Pourmand; Rahil Mashhadi; Kiarash Ghazvini; Epidemiology of efflux pumps genes mediating resistance among Staphylococcus aureus; A systematic review.. Microbial Pathogenesis 2020, 139, 103850, 10.1016/j.micpath.2019.103850.
- Hasan Ghajavand; Mansour Kargarpour Kamakoli; Sharareh Khanipour; Shahin Pourazar Dizaji; Morteza Masoumi; Fatemeh Rahimi Jamnani; Abolfazl Fateh; Mehdi Yaseri; Seyed Davar Siadat; Farzam Vaziri; et al. Scrutinizing the drug resistance mechanism of multi- and extensively-drug resistant Mycobacterium tuberculosis: mutations versus efflux pumps. Antimicrobial Resistance & Infection Control 2019, 8, 70, 10.1186/s13756-019-0516-4.
- Sravan Kumar Miryala; Anand Anbarasu; Sudha Ramaiah; Systems biology studies in Pseudomonas aeruginosa PA01 to understand their role in biofilm formation and multidrug efflux pumps. Microbial Pathogenesis 2019, 136, 103668, 10.1016/j.micpath.2019.103668.
- Bruno Casciaro; Debarun Dutta; Maria Rosa Loffredo; Stefania Marcheggiani; Alison M McDermott; Mark Willcox; Maria Luisa Mangoni; Esculentin‐1a derived peptides kill Pseudomonas aeruginosa biofilm on soft contact lenses and retain antibacterial activity upon immobilization to the lens surface. Peptide Science 2018, 110, e23074, 10.1002/bip.23074.
- Marc Crouzet; Stephane Claverol; Anne-Marie Lomenech; Caroline Le Senechal; Patricia Costaglioli; Christophe Barthe; Bertrand Garbay; Marc Bonneu; Sébastien Vilain; Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLOS ONE 2017, 12, e0180341, 10.1371/journal.pone.0180341.
- Weslley Felix De Oliveira; P.M.S. Silva; R.C.S. Silva; G.M.M. Silva; G. Machado; L.C.B.B. Coelho; M.T.S. Correia; Staphylococcus aureus and Staphylococcus epidermidis infections on implants. Journal of Hospital Infection 2018, 98, 111-117, 10.1016/j.jhin.2017.11.008.
- Bruno Casciaro; María Moros; Sara Rivera-Fernández; Andrea Bellelli; Jesús M. De La Fuente; Maria Luisa Mangoni; Gold-nanoparticles coated with the antimicrobial peptide esculentin-1a(1-21)NH 2 as a reliable strategy for antipseudomonal drugs. Acta Biomaterialia 2017, 47, 170-181, 10.1016/j.actbio.2016.09.041.
- Ivana D'angelo; Bruno Casciaro; Agnese Miro; Fabiana Quaglia; Maria Luisa Mangoni; Francesca Ungaro; Overcoming barriers in Pseudomonas aeruginosa lung infections: Engineered nanoparticles for local delivery of a cationic antimicrobial peptide. Colloids and Surfaces B: Biointerfaces 2015, 135, 717-725, 10.1016/j.colsurfb.2015.08.027.
- Bruno Casciaro; Ivana D’Angelo; Xiaoping Zhang; Maria Rosa Loffredo; Gemma Conte; Floriana Cappiello; Fabiana Quaglia; Yuan-Pu Peter Di; Francesca Ungaro; Maria Luisa Mangoni; et al. Poly(lactide-co-glycolide) Nanoparticles for Prolonged Therapeutic Efficacy of Esculentin-1a-Derived Antimicrobial Peptides against Pseudomonas aeruginosa Lung Infection: in Vitro and in Vivo Studies. Biomacromolecules 2019, 20, 1876-1888, 10.1021/acs.biomac.8b01829.
- Virginio Cepas Lopez; Sara M. Soto; The Usefulness of Microalgae Compounds for Preventing Biofilm Infections. Antibiotics 2019, 9, 9, 10.3390/antibiotics9010009.
- Bruno Casciaro; Qiao Lin; Sergii Afonin; Maria Rosa Loffredo; V. De Turris; Volker Middel; Anne S. Ulrich; Yuanpu Peter Di; Maria Luisa Mangoni; Inhibition of Pseudomonas aeruginosa biofilm formation and expression of virulence genes by selective epimerization in the peptide Esculentin‐1a(1‐21) NH 2. The FEBS Journal 2019, 286, 3874-3891, 10.1111/febs.14940.




