
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ibrahimm Mugerwa | + 2767 word(s) | 2767 | 2021-10-13 12:18:04 | | | |
| 2 | Lindsay Dong | + 124 word(s) | 2891 | 2021-11-25 04:08:29 | | |
Video Upload Options
Uganda is making strides and progress with regard to developing and implementing a functional AMR surveillance strategy for human health. Although numerous challenges still exist, following the laboratory health system strengthening approach, the readily addressable issues are with the health infrastructure, its integration, capacity building and operation. Antibiotic resistance and its mechanisms have now been in existance for over decades, and its drivers in both clinical, human, agriculture-veterinary go beyond the community and clinical aspects. Its noteworthy that the emergency of resistance is a natural phenomenon in the environment but kin to climate change.
If left unchecked, this has great potential of undoing all medical and agricultural advancements of the entire previous century. Implementation of country driven program based surveillance that embraces a One Health Approach is the ideal approach to understanding and solving this one health challenge that seems to be wicked in solve at the face mankind.
1. Introduction
Globally, bacterial infections cause acute and chronic life-threatening infections and are becoming difficult to treat due to the increasing emergence and spread of antimicrobial resistance. We have known about resistance and its underlying mechanism for close to seven decades, according to [1] but efforts to tackle resistance in routine practice are only starting to gain momentum. The drivers and determinants of antimicrobial resistance are complex and cut across strata of life and ecosystems, i.e., microbial, niche, food systems, human health and the environment in general [2].
Indeed, antimicrobial resistance is a natural phenomenon; however, its rapidly increasing prevalence is driven by misuse and overuse of antibiotics [3][4]. If not addressed, antimicrobial resistance will cost the global economy over 100 trillion USD by 2050 with a projection of over 10 million death annually [5]. To this end, AMR surveillance is considered a high-yield investment that countries must undertake to mitigate this global threat. Through surveillance, countries can monitor trends and detect the magnitude of the emerging resistance to priority antibiotics. This evidence can inform local policies both at the national and sub-national level in areas of treatment guidelines, as well as infection prevention and control strategies.
The World Health Organisation, through its global Antimicrobial Resistance Surveillance System (GLASS), provides baseline guidelines for surveillance, and most national action plans share the ambitions set out in this document. The guidelines serve as a framework to safeguard the integrity of antimicrobials whose utility is critical for infection management. For example, Uganda’s national action plan (NAP) aims to prevent, slow down and control the spread of resistant pathogens as indicated on http://www.cphl.go.ug/policy-documents (accessed on 18 August 2021). A robust surveillance system is integral to achieving these aims. Since its publication in 2018, various stakeholders along the AMR chain (human, animal health and environment) have been involved in developing and implementing components of the NAP.
2. Design of Sentinel Sites
2.1. Establishment of AMR Governance Structures at National and Sub-National Level
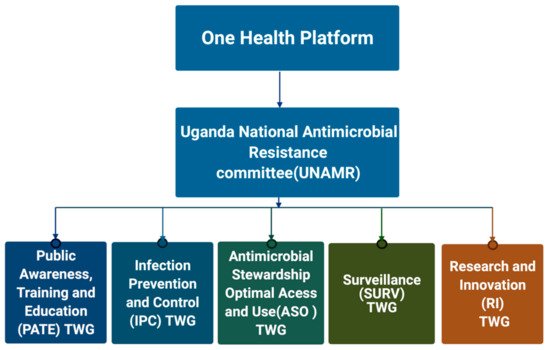
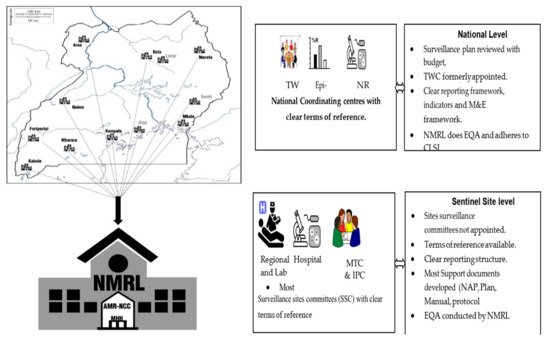
2.2. Human Health AMR Laboratory Diagnostics
Sample collection is designed to reflect the referral system of the healthcare system in Uganda, which in itself allows for examining the changes in AMR profiles as patients are escalated through the referral system. Since patients spend longer periods in hospital at higher strata of the referral system, we expect that the odds of exposure to hospital-generated AMR increase and so will the detectable resistance [7]. The location of sentinel sites is critical as it has a direct impact on the time of sample collection to processing. This in turn has an effect on the recovery of microbes for AMR susceptibility profiling in this analysis.
2.3. Integration and Development of Linkages for Electronic Data Interoperability and Sharing
Uganda has leveraged existing Ministry of Health laboratory information platforms built on the health laboratory infrastructure to form a laboratory information system known as the African Laboratory Information System (ALIS). This information system has been developed as a generic tool for the country’s laboratory system and programmed to integrate with other systems and tools such as WHO-NET. The system takes all the key variables on the patient laboratory request form so that patient-level data can be analysed to generate epidemiological reports automatically.
2.4. AMR Data Management at Facility and Reference Laboratory Level{ TC “AMR Data Management at Facility and Reference Laboratory Level” \f C \l “1” }
At the sub-national facility level, in the microbiology laboratory request forms and the daily activity registers, patient level data is entered into WHONET, a software application that analyses these data. Working with the facility implementing partner, data entry is validated at the facility level prior to being shared with the National Coordination Centre. Data validation at the national level is performed by the Technical Working Committee on surveillance that scrutinizes the data prior to external uploading onto the WHO-GLASS. SOPs and developed guidelines for data collection, sampling and reporting in surveillance sites are standardized: strengthen data management at AMR secretariat, NCC and sentinel sites by delivering IT infrastructure, paper-based forms and registers to sites, e.g., microbiology referral forms and registers; and train laboratorians to analyse microbiology lab AST data. Anonymized isolate level data are submitted to the AMR-NCC through an electronic system at the national level, and this will eventually be linked to the national database (DISC) for evaluation of quality. Monthly AMR and microbiology data are reported to DHIS2.
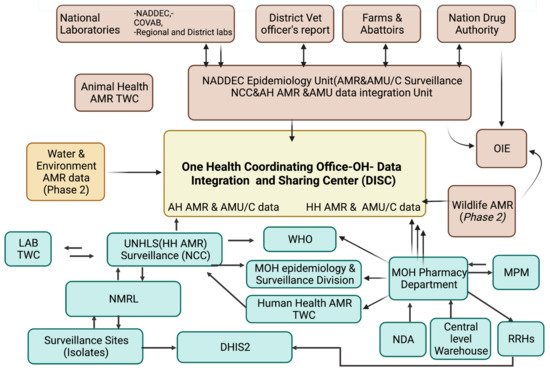
2.5. Surveillance for AMR Stewardship Optimal Use, Access and Consumption { TC “Surveillance for AMR Stewardship Optimal Use, Access and Consumption” \f C \l “1” }
As guided by the AMR-NAP, the development of a successful national AMU/C surveillance program is on-going currently with the process of policy and guideline development, and reviews as pre-requisite to ministry approval for operationalization. The protocols and guidelines are currently being drafted and presented to the ministries as well as the One Health technical working committees to guide and update the surveillance plans.
2.6. Approaches for AMR Data Management in Human Health{ TC “10.0 METHODS AND APPROACHES FOR AMR DATA MANAGEMENT IN HUMAN HEALTH” \f C \l “1” }
AMR surveillance in low- and middle-income settings [8] reflects Uganda’s perspective. Raw data on 10 national priority pathogens classified for country surveillance were collected following a recent study of acute febrile illness (AFI) alongside the clinical samples received by these laboratories for routine patient care, [9]. As an initial step, Uganda designated sentinel sites to perform AFI surveillance studies with the support of the CDC and the Global Health Security Agenda. The project and its data helped to inform the initiation of the national surveillance program as a basis to select sites with minimal capacity. It was this first set of data that was used in the first country AMR WHO-GLASS report.
Following a routine case-based surveillance approach towards priority specimens (blood, urine, stool, genital swabs and CSF), these samples are sent to microbiology laboratories for the purposes of clinical testing at the four sentinel sites. Here, using patient level data tools, epidemiological, clinical and demographic data were also collected alongside the microbiology data. After basic microbiology cultures and sensitivity testing, isolates are sent to the National Microbiology Reference Laboratory (NMRL) for validation and archiving. At the end of every quarter of a year, the NMRL must conduct site visits for retrospective on-site data cleaning and support supervision for improved microbiology culture and sensitivity laboratory procedures.
2.7. Processing and Quality Control for AMR Data{ TC “Process Quality Control for AMR Data” \f C \l “1” }
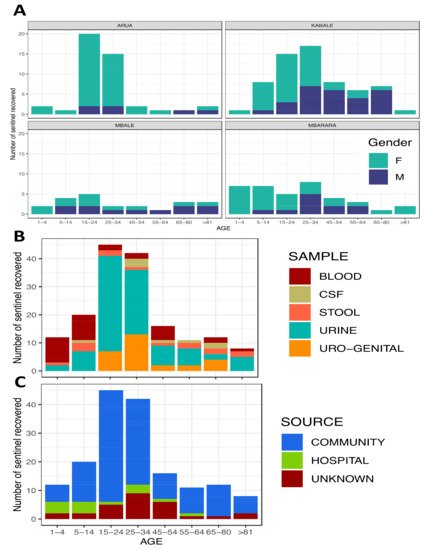
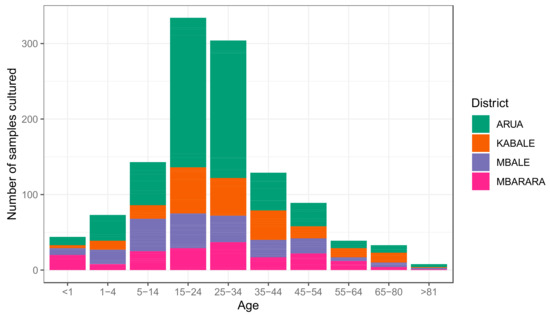
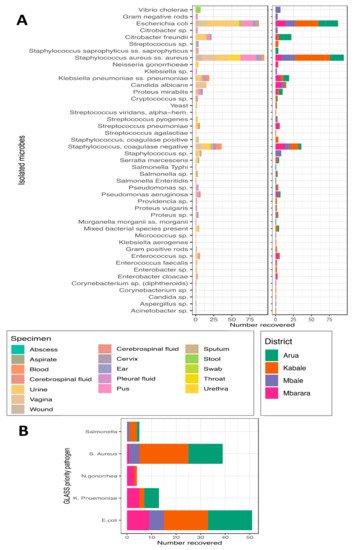
3. Preliminary Results and Outputs
Preliminary Antibiotic Resistance Profiles
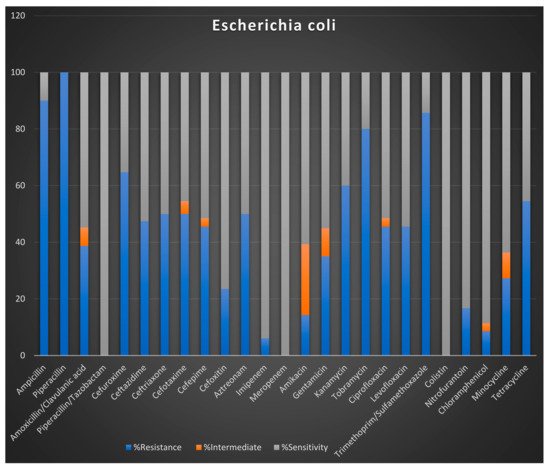
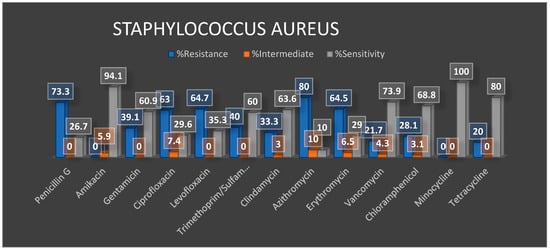
4. Summary
References
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433.
- Gelband, H.; Molly Miller, P.; Pant, S.; Gandra, S.; Levinson, J.; Barter, D.; White, A.; Laxminarayan, R. The state of the world’s antibiotics. Wound Health S. Afr. 2015, 8, 30–34.
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. Rev. Antimicrob. Res. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 14 September 2021).
- Alhomoud, F.; Almahasnah, R.; Alhomoud, F.K. “You could lose when you misuse”–factors affecting over-the-counter sale of antibiotics in community pharmacies in Saudi Arabia: A qualitative study. BMC Health Serv. Res. 2018, 18, 1–9.
- De Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184.
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS): Technical Meeting on the Early Implementation Phase: 22–23 October 2015: WHO Regional Office for Europe Copenhagen, Denmark: Meeting Report (No. WHO/OHE/PED/AMR/2016.1). 2016. Available online: https://apps.who.int/iris/bitstream/handle/10665/259744/9789241513449-eng.pdf (accessed on 6 September 2021).
- Peters, L.; Olson, L.; Khu, D.T.; Linnros, S.; Le, N.K.; Hanberger, H.; Hoang, N.T.B.; Tran, D.M.; Larsson, M. Multiple antibiotic resistance as a risk factor for mortality and prolonged hospital stay: A cohort study among neonatal intensive care patients with hospital-acquired infections caused by gram-negative bacteria in Vietnam. PLoS ONE 2019, 14, e0215666.
- Seale, A.C.; Gordon, N.C.; Islam, J.; Peacock, S.J.; Scott, J.A.G. AMR Surveillance in low and middle-income settings‒A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res. 2017, 26, 1–17.
- Kibuuka, A.; Byakika-Kibwika, P.; Achan, J.; Yeka, A.; Nalyazi, J.N.; Mpimbaza, A.; Rosenthal, P.J.; Kamya, M.R. Bacteremia Among Febrile Ugandan Children Treated with Antimalarials Despite a Negative Malaria Test. Am. J. Trop. Med. Hyg. 2015, 93, 276–280.
- Lugsomya, K.; Chatsuwan, T.; Niyomtham, W.; Tummaruk, P.; Hampson, D.J.; Prapasarakul, N. Routine prophylactic antimicrobial use is associated with increased phenotypic and genotypic resistance in commensal Escherichia coli isolates recovered from healthy fattening pigs on farms in Thailand. Microb. Drug Resist. 2018, 24, 213–223.
- Pungcharoenkijkul, S.; Traipattanakul, J.; Thunyaharn, S.; Santimaleeworagun, W. Antimicrobials as Single and Combination Therapy for Colistin-Resistant Pseudomonas aeruginosa at a University Hospital in Thailand. Antibiotics 2020, 9, 475.
- Uganda Essential Medicines List. 2016. Available online: https://www.health.go.ug/ (accessed on 18 August 2021).
- Devi, L.S.; Broor, S.; Rautela, R.S.; Grover, S.S.; Chakravarti, A.; Chattopadhya, D. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: A 3-year study. Int. J. Appl. Basic Med. Res. 2020, 10, 156–163.




