Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jonathan A.G. Cox | + 2442 word(s) | 2442 | 2021-11-15 04:54:52 | | | |
| 2 | Rita Xu | Meta information modification | 2442 | 2021-11-15 12:24:49 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Cox, J.A. Microfluidics as a Novel Technique for Tuberculosis. Encyclopedia. Available online: https://encyclopedia.pub/entry/16004 (accessed on 08 February 2026).
Cox JA. Microfluidics as a Novel Technique for Tuberculosis. Encyclopedia. Available at: https://encyclopedia.pub/entry/16004. Accessed February 08, 2026.
Cox, Jonathan A.g.. "Microfluidics as a Novel Technique for Tuberculosis" Encyclopedia, https://encyclopedia.pub/entry/16004 (accessed February 08, 2026).
Cox, J.A. (2021, November 15). Microfluidics as a Novel Technique for Tuberculosis. In Encyclopedia. https://encyclopedia.pub/entry/16004
Cox, Jonathan A.g.. "Microfluidics as a Novel Technique for Tuberculosis." Encyclopedia. Web. 15 November, 2021.
Copy Citation
Tuberculosis (TB) remains a global healthcare crisis, with an estimated 5.8 million new cases and 1.5 million deaths in 2020. TB is caused by infection with the major human pathogen Mycobacterium tuberculosis, which is difficult to rapidly diagnose and treat.
tuberculosis
Mycobacterium
diagnostics
drug discovery
antibiotics
1. Introduction
1.1. Tuberculosis and Its Global Health Threat
Mycobacterium tuberculosis is the causative agent of the human pulmonary infection tuberculosis (TB). TB is the second leading infectious killer since the global COVID-19 pandemic. The World Health Organisation (WHO) estimates that 1.5 million people died from TB in 2020 [1]. Despite most TB strains being treatable with antibiotics, some of the key medical challenges include achieving rapid diagnostics, the rise of multidrug-resistant TB, and the poor treatment efficacy of latent TB. The current recommended treatment for drug-susceptible TB takes a minimum six-month administration of isoniazid (INH), rifampicin (RIF), pyrazinamide (PZA), and ethambutol (EMB) [2]. This first-line recommendation has failed to adapt in the last thirty-five years despite the increasing occurrence of drug resistance. Recently, a phase 3 trial provided evidence of a four-month treatment regimen with rifapentine and moxifloxacin [3]. Additionally, many efforts have been made to reduce the mycobacterial burden (reducing mortality and transmission), eradicate persistent mycobacterial populations, and to reduce drug resistance through various incentives such as END-TB [4] and WHO End TB Strategy 2016–2035 [5]. Research into the economic burden of TB has revealed a global cost of 983 bn USD from 2015–2030 if the current health status continues [6]. There is a pressing need for innovative advancements and applications which combine multidisciplinary research for combating the looming crisis of TB.
1.2. Mycobacterium Tuberculosis and the Pathogenesis of TB
M. tuberculosis is a rod-shaped acid-fast-staining bacterium of the Actinomycete family [7]. The unique “waxy” cell envelope of M. tuberculosis contains a core composed of peptidoglycan and the highly branched polysaccharide arabinogalactan. This is covalently attached to the unique mycolic acids that cover the bacteria with a mycobacterial outer membrane which allows cellular integrity and virulence [8]. This self-protection permits the organism to evade the host immune system and prevents antibiotic penetration [8][9]. The molecular pathology by which M. tuberculosis evades the host and causes disease is complex, involving a dynamic range of immune cells. The organism infects the host after the inhalation of droplet nuclei spread by aerosolisation from an infected individual, which then resides in the respiratory tract [10]. There are various types of infection that can manifest from M. tuberculosis in individuals—one where the infection clears, one with an active infection treated with a course of antibiotics, and one which remains in a latent form [11]. Upon infection, the early innate immune system emerges with an influx of neutrophils, monocytes, macrophages, and dendritic cells of the lungs [12]. Through phagocytosis, bacteria are consumed by alveolar macrophages to form a phagosome and then subsequently eliminated through the formation of phagolysosomes [13]. However, M. tuberculosis can avoid this host defence response by persisting in phagosomes and inhibiting lysosome fusion [13]. The subsequent established intracellular infection and influx of immune cells which surround the site of infection forms a tuberculous granuloma [14]. The early granuloma (Figure 1) consists of the infected macrophages in the centre, enclosed by foamy macrophages and other mononucleated cells, and surrounded by lymphocytes [15]. During the maturation of the granuloma, a fibrous capsule encloses the macrophage centre and eventually forms necrotic lesions, leading to caseation [14][15].
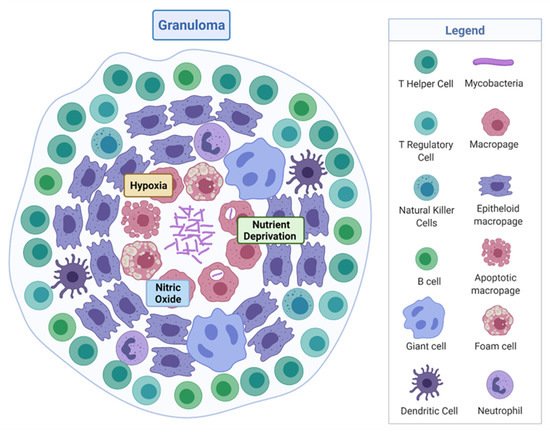
Figure 1. Tuberculous granuloma. Encapsulated Mycobacterium tuberculosis surrounded by immune cells, creating a hypoxic, nutrient-deprived, and nitric oxide environment. Adapted from “Granuloma”, by BioRender.com (2020). Retrieved from https://app.biorender.com/biorender-templates, accessed on 27 September 2021.
Here, M. tuberculosis can survive in a dormancy state known as non-replicating persistence (NRP). The external pressures such as hypoxia, nutrient deprivation, low pH, and high CO2 created by the hostile host environment induce this survival response of the bacteria [16]. The NRP state can relapse into active disease, especially in high-risk groups such as immunodeficient individuals, persons infected with human immunodeficiency virus or undergoing organ and haematologic transplantations [11]. Houben and Dodd (2016) previously estimated that NRP TB infected approximately 1.7 billion people in 2014 by generating an annual risk model of infection between 1934 and 2014 [17]. The issue posed by the ability of NRP M. tuberculosis to effectively hide within the hostile environment of the granuloma is that not only does the immune system keep the bacteria trapped, it also physically restricts penetration by antimicrobials, thus protecting M. tuberculosis from antibiotic activity.
1.3. Current Diagnostics, Research Methods, and Treatment
Early diagnosis and accurate detection of TB infection is essential for effective treatment options, especially in low-income and high-burden countries. Conventional TB diagnostics include microscopy (Ziehl–Neelsen staining), which provides 22–43% low sensitivity for a single smear [18]. Other methods include chest radiography, which is limited in resource-constraint locations [19], and liquid/solid culturing, which requires suitable levels of biosafety [20]. Diagnosis of latent infection requires a tuberculin skin test or interferon-gamma release assays. However, both of these tests do not identify individuals that will progress to active disease [20].
The phenotypic evaluation of clinical isolates, by culturing M. tuberculosis in the laboratory in the presence of different concentrations of antimicrobials, is traditionally used to detect drug-resistant strains. The turnover time for these results is extensive, by which point the patient’s health will have deteriorated [21]. Improvements in molecular diagnostic testing have revolutionised detection, such as the genotypic test Cepheid GeneXpert MTB/RIF, which can give a readout in two hours of TB detection and RIF resistance [22]. Additionally, whole-genome sequencing of TB is expanding with support from the WHO but still relies on culturing samples for weeks and technical methods in preparing genomic DNA for sequencing [23]. User-friendly and non-laborious detection methods, which are portable, are required to improve detection time at lower cost.
Experimental modelling of TB has historically helped scientists to discover the pathogenicity, physiology, metabolism, and genetic make-up of the organism. Challenges arising for researchers studying mycobacteria are the characteristics of slow growth rate, hydrophobic aggregation of cells in the absence of non-ionic surfactant when grown in culture, and the need for the containment of aerosols which brings additional safety precautions, including a biosafety level 3 (BSL-3) facility [7]. Additionally, the investigation of heterogenicity is difficult in bulk cultures compared with single-cell analysis [24]. Animal models are abundant for studying TB, such as zebrafish, rabbits, guinea pigs, and mouse models [25]. However, absent is the ability of each model to represent all aspects of the physiological state of the cell and tissue environment [25], or they lack the lung immune system entirely [26]. There have been extensive reviews detailing the methods used to experimentally model this organism in its non-replicating state [27][28][29]. However, to date, no NRP models mimic all the physiological features of the bacteria in this condition. Therefore, novel in vitro experimental models of TB are imperative.
Research groups often use variable types of nutrient media, inoculum starting points, and reading endpoints, making the standardisation of antimicrobial testing for M. tuberculosis difficult. Efforts have been made to standardise testing; however, protocols are still time consuming [30][31]. TB has shown resistance to antimicrobials, including multidrug-resistant strains resistant to RIF and INH [1]. Worryingly, extensively drug-resistant TB is increasing, which is resistant to RIF, INH, Fluoroquinolone, and Kanamycin [32].
2. Microfluidics
2.1. Technological Advancement of Microfluidics
We require innovative and advanced technology to advance M. tuberculosis research, such as new high-throughput methods of phenotypic assessment. Advancements in microtechnology, particularly at the micro and nanoscale, have had wide microbiological applications. Microfluidics is a rapidly growing field which comprises multidisciplinary expertise in biology, chemistry, physics, and engineering. A simple definition of microfluidics is the systematic manipulation of systems that have microscale channels where fluid volumes of nanolitres to attolitres can flow in geometric configurations [33]. Well suited to the scale of bacteria, microfluidics can produce biological assays in parallel with well-defined, controllable environmental conditions. Advantageously, the methodological approach of manipulating fluids opens a pathway to reduce animal models. This review will outline the field of microfluidics, and discuss the recent use of microfluidic techniques in TB diagnostics and drug discovery.
2.2. Droplet Microfluidics
The study of multiphase flows, often termed droplet microfluidics, is a subset of the microfluidics field in which nano- to femtoliter volume droplets can be routinely generated by drop-making micronozzles in a carrier fluid at production rates exceeding 10 kHz (Figure 2). Recently, droplet production rates exceeding 1 MHz have been reported [34]. High droplet production rates enable the possibility of undertaking millions of individual experiments within a single microfluidic device. Further, droplet microfluidic systems enable the efficient control of droplet volumes, repeatable and reliable droplet manipulation, high-throughput capability, single-cell analysis capabilities, and can be fully automated. Their applications include chemical and biological assays [35][36], inorganic chemistry [37], and protein crystallisation [38][39]. The reader is referred to recent reviews on various assays, screens, and studies enabled by droplet microfluidics [40], as well as its applications in drug discovery, transcriptomics, and molecular genetics [41].
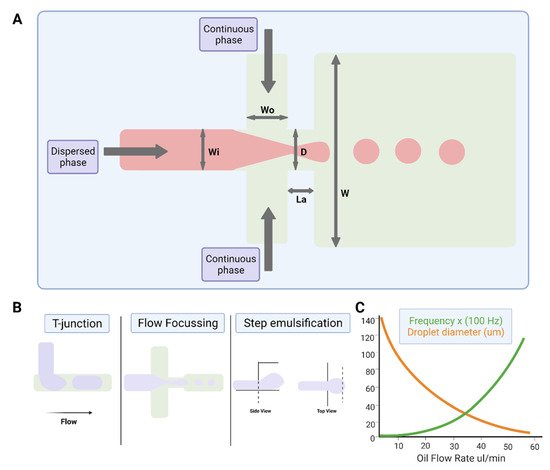
Figure 2. Water-in-oil droplet-generation microfluidics. (A) Production of water-in-oil droplets using a flow-focusing design. The dispersed phase is squeezed by two counter-streaming flows of the carrier phase, forcing drops to form and detach. (B) Droplet generation using T-junction, flow-focusing geometry and step emulsification. (C) Graph showing that droplet size decreases, and frequency of formation increases with increasing oil flow rate. Figures created on Biorender.com, accessed on 9 November 2021.
3. Recent Application of Microfluidics for TB
Applications of Microfluidics for Diagnostics and Detecting Drug-Resistant Strains
Employing microfluidics is a promising approach for rapid and cost-effective diagnostics for M. tuberculosis. Detecting the pathogen with robust and reproducible fluidic models offers capabilities for clinical procedures and scientific exploration. Interestingly, a bacteria enrichment microfluidic chip and a microfluidic immunoassay chip have detected airborne M. tuberculosis. Jing and colleagues (2014) validated a method whereby a micro-pump draws air containing bacteria into the enrichment fluidic chip and then a full immunoassay reaction is performed on a separate chip. The method offers the potential to accurately screen M. tuberculosis in the aerosol [42]. Airborne M. tuberculosis currently requires long cultivation due to the low concentration in air samples. Capturing and directly detecting airborne M. tuberculosis will aid effective disease prevention and control as there is a requirement to detect samples directly from patients for quicker analysis. The small volume sizes in microfluidic chip cultivation provides rapid detection at lower sample concentrations. Diagnosing TB, especially in developing countries, requires low-cost point-of-care technologies. A paper-based microfluidics system detected sputum samples containing mycobacteria. The system used enabled the decontamination of non-mycobacteria and storage of the sputum sample [43]. A laser-etched indium tin oxide glass and PDMS microfluidic chip were used to rapidly detect and quantitate M. tuberculosis with high sensitivity within forty-five minutes. By creating an eight-chamber microfluidic electrochemical system with real-time loop-mediated isothermal amplification (LAMP), amplification of three respiratory related infections including M. tuberculosis could be monitored by measuring the electrochemical signal of methylene blue [44]. Here, a microfluidic chip, with different sample chambers, provides cost- and time-efficient detection which would benefit clinicians to decide on optimal antibiotic treatments.
Six species of mycobacteria, including nontuberculous species and members of the M. tuberculosis complex, were detected by combining a closed system of bead-beating, droplet fluidics, and surface-enhanced Raman spectroscopy. The spectral information obtained from the vibrational signals of the mycobacterial cell wall component, mycolic acid, effectively identified the different species. This is a promising step forward for ensuring the correct treatments are administered for the correct infections [45]. Small channel dimensions enable the manipulation of cell environments and thus can represent improved biological investigation. A potential method for quantitively detecting M. tuberculosis in droplet microfluidics was developed by detecting cells that express the endogenous β-lactamase, BlaC—an enzyme marker naturally expressed by M. tuberculosis. By encapsulating a specific fluorescent probe of BlaC and samples of bacterial strains that express BlaC in droplets, the researchers could calculate the initial concentration of cells based on fluorescence [46].
Other researchers have combined PCR techniques with microfluidics. Ip et al. (2018) used a single chip comprising positive and negative reaction chambers, as well as small liquid handling chambers. They performed isolation of M. tuberculosis H37Ra with magnetic beads and differentiation of live/dead bacteria with propidium monoazide dye, followed by RT-PCR and optical detection within two hours. By measuring the threshold cycle number, a low detection limit of 14 colony-forming units per reaction was achieved [47]. Besides the above new PCR microfluidic approaches, genetically detecting M. tuberculosis without the laborious need for PCR amplification has been achieved. For example, Domínguez et al. (2015) created a micro-cantilever platform, where hydration-induced stress could identify M. tuberculosis and RIF resistance within 1.5 h [48].
Previously, label-free DNA of M. tuberculosis from clinical isolates was detected by an integrated system of microfluidics and electrochemical biosensing. The platform consisting of a monolithic chip and multiwall carbon nanotubes detects M. tuberculosis without the need for DNA amplification [49]. Another biosensing device was developed to detect MPT64—an antigen secreted by M. tuberculosis. The protein is a biomarker for actively dividing mycobacteria, detected by electrochemical impedance spectroscopy and synthetic aptamers integrated with a microfluidic chamber [50].
Detecting drug-resistant strains early in the infection will aid clinical decision making and shorten the time for optimal drug treatment. Sophisticated detection of resistant strains will also transform drug discovery and innovation within the laboratory. Researchers detected single-nucleotide polymorphisms between RIF-resistant M. tuberculosis isolates and susceptible isolates by combining a microfluidic chip with post-PCR high-resolution melting analysis (HRMA). The authors’ “Light Forge” microfluidic DNA melting-based TB test showed better performance of melting temperature differences compared to conventional Sanger sequencing, as well as a HRMA device on its own and phenotypic drug susceptibility testing [51]. Additionally, evidence shows that by incorporating open-chip microfluidics with padlock probe (PLP) ligation and rolling circle amplification (RCA), a two-hour assay is achievable for detecting an INH resistance caused by mutations in the gene (katG) in M. tuberculosis. The lab-on-a-disc platform utilised separate fluidic chambers for ligation and amplification steps, which provided temperature control [52]. Law et al. (2018) combined a lab-on-a-disk and recombinase polymerase amplification to fluorescently detect the pathogen with a sensitivity of 102 colony-forming units per millilitre [53]. Drug-resistant strains to β-lactams were fluorescently detected using a droplet-based microfluidic device and a custom 3D particle counter (Figure 3). The microfluidic chip comprised separate input channels for bacteria, ampicillin and broth mixture, fluorocillin (a β-lactamase sensor), and oil to encapsulate single bacteria cells into droplets. Antibiotic-resistant clinical isolates could grow inside the droplets, detected by fluorescent microscopy [54].
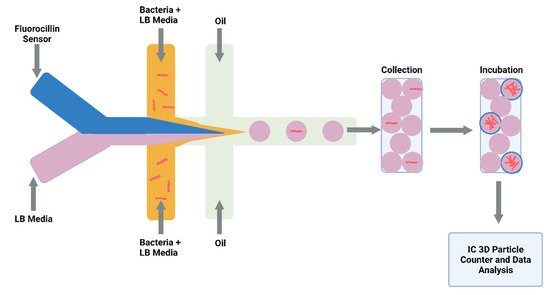
Figure 3. Schematic illustration of droplet system coupled to an Integrated Comprehensive Droplet Digital Detection. Flow-focusing microfluidic chip geometry producing encapsulated mycobacteria in droplets. Figure created on Biorender.com, accessed on 26 October 2021.
Investigators are overcoming the challenge of genotyping drug-resistant strains of the pathogen directly from sputum. Researchers detected and genotyped RIF and INH resistance by creating a closed system composed of a microfluidic amplification microarray [55]. Likewise, the lab-on-a-film platform created by Kukhtin et al. (2020) integrated amplification, hybridisation, washing, and imaging. The authors reported M. tuberculosis detection in sputum as 43 CFU/mL; however, future work of this method includes sensitivity investigation [56].
References
- World Health Organization. Global Tuberculosis Report 2021; World Health Organization: Albany, NY, USA, 2021.
- Lee, S.H. Tuberculosis Infection and Latent Tuberculosis. Tuberc. Respir. Dis. 2016, 79, 201–206.
- Dorman, S.E.; Nahid, P.; Kurbatova, E.V.; Phillips, P.P.J.; Bryant, K.; Dooley, K.E.; Engle, M.; Goldberg, S.V.; Phan, H.T.T.; Hakim, J.; et al. Four-Month Rifapentine Regimens with or without Moxifloxacin for Tuberculosis. N. Engl. J. Med. 2021, 384, 1705–1718.
- END-TB. Expand New Drug Markets for TB; END-TB 2021; 2016; pp. 1–30. Available online: https://unitaid.org/assets/Swiss-TPH-endTB-MTE-final-report-rvd-final-13-Jun-18.pdf (accessed on 26 October 2021).
- World Health Organization. The END-TB Strategy; World Health Organization: Albany, NY, USA, 2012.
- Burki, T.K. The global cost of tuberculosis. Lancet Respir. Med. 2018, 6, 13.
- Parish, T.; Stoker, N.G. Mycobacteria: Bugs and bugbears (Two steps forward and one step back). Mol. Biotechnol. 1999, 13, 191–200.
- Batt, S.M.; Minnikin, D.E.; Besra, G.S. The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem. J. 2020, 477, 1983–2006.
- Minnikin, D.E.; Kremer, L.; Dover, L.G.; Besra, G.S. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 2002, 9, 545–553.
- Jee, B. Understanding the early host immune response against Mycobacterium tuberculosis. Cent. Eur. J. Immunol. 2020, 45, 99–103.
- Getahun, H.; Matteelli, A.; Chaisson, R.E.; Raviglione, M. Latent Mycobacterium tuberculosis infection. N. Engl. J. Med. 2015, 372, 2127–2135.
- Ernst, J.D. The immunological life cycle of tuberculosis. Nat. Rev. Immunol. 2012, 12, 581–591.
- Pieters, J. Entry and survival of pathogenic mycobacteria in macrophages. Microbes Infect. 2001, 3, 249–255.
- Ramakrishnan, L. Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 2012, 12, 352–366.
- Russell, D.G.; Cardona, P.-J.; Kim, M.-J.; Allain, S.; Altare, F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009, 10, 943–948.
- Martin, C.J.; Carey, A.F.; Fortune, S.M. A bug’s life in the granuloma. Semin. Immunopathol. 2016, 38, 213–220.
- Houben, R.M.G.J.; Dodd, P.J. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Med. 2016, 13, e1002152.
- Singhal, R.; Myneedu, V.P. Microscopy as a diagnostic tool in pulmonary tuberculosis. Int. J. Mycobacteriol. 2015, 4, 1–6.
- Walzl, G.; McNerney, R.; du Plessis, N.; Bates, M.; McHugh, T.D.; Chegou, N.N.; Zumla, A. Tuberculosis: Advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis 2018, 18, e199–e210.
- Pai, M.; Nicol, M.P.; Boehme, C.C. Tuberculosis Diagnostics: State of the Art and Future Directions. Microbiol. Spectr. 2016, 4, 4–5.
- Lee, A.; Xie, Y.L.; Barry, C.E.; Chen, R.Y. Current and future treatments for tuberculosis. BMJ 2020, 368, m216.
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 2010, 363, 1005–1015.
- Meehan, C.J.; Goig, G.A.; Kohl, T.A.; Verboven, L.; Dippenaar, A.; Ezewudo, M.; Farhat, M.R.; Guthrie, J.L.; Laukens, K.; Miotto, P.; et al. Whole genome sequencing of Mycobacterium tuberculosis: Current standards and open issues. Nat. Rev. Microbiol. 2019, 17, 533–545.
- Toniolo, C.; Rutschmann, O.; McKinney, J.D. Do chance encounters between heterogeneous cells shape the outcome of tuberculosis infections? Curr. Opin. Microbiol. 2021, 59, 72–78.
- Singh, A.K.; Gupta, U.D. Animal models of tuberculosis: Lesson learnt. Indian J. Med. Res. 2018, 147, 456–463.
- Rhoades, E.R.; Frank, A.A.; Orme, I.M. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. Tuber. Lung Dis. 1997, 78, 57–66.
- Gibson, S.E.R.; Harrison, J.; Cox, J.A.G. Modelling a Silent Epidemic: A Review of the In Vitro Models of Latent Tuberculosis. Pathogens 2018, 7, 88.
- Parish, T. In vitro drug discovery models for Mycobacterium tuberculosis relevant for host infection. Expert Opin. Drug Discov. 2020, 15, 349–358.
- Batyrshina, Y.R.; Schwartz, Y.S. Modeling of Mycobacterium tuberculosis dormancy in bacterial cultures. Tuberculosis 2019, 117, 7–17.
- Schön, T.; Werngren, J.; Machado, D.; Borroni, E.; Wijkander, M.; Lina, G.; Mouton, J.; Matuschek, E.; Kahlmeter, G.; Giske, C.; et al. Antimicrobial susceptibility testing of Mycobacterium tuberculosis complex isolates—The EUCAST broth microdilution reference method for MIC determination. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1488–1492.
- Kim, S.J. Drug-susceptibility testing in tuberculosis: Methods and reliability of results. Eur. Respir. J. 2005, 25, 564–569.
- Centers for Disease Control and Prevention. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs—Worldwide, 2000–2004. MMWR. Morb. Mortal. Wkly. Rep. 2006, 55, 301–305.
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373.
- Shim, J.U.; Ranasinghe, R.T.; Smith, C.A.; Ibrahim, S.M.; Hollfelder, F.; Huck, W.T.; Klenerman, D.; Abell, C. Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays. ACS Nano 2013, 7, 5955–5964.
- Vyawahare, S.; Griffiths, A.D.; Merten, C.A. Miniaturization and parallelization of biological and chemical assays in microfluidic devices. Chem. Biol. 2010, 17, 1052–1065.
- Theberge, A.B.; Courtois, F.; Schaerli, Y.; Fischlechner, M.; Abell, C.; Hollfelder, F.; Huck, W.T.S. Microdroplets in Microfluidics: An Evolving Platform for Discoveries in Chemistry and Biology. Angew. Chem. Int. Ed. 2010, 49, 5846–5868.
- Abou-Hassan, A.; Sandre, O.; Cabuil, V. Microfluidics in Inorganic Chemistry. Angew. Chem. Int. Ed. 2010, 49, 6268–6286.
- Zheng, B.; Gerdts, C.J.; Ismagilov, R.F. Using nanoliter plugs in microfluidics to facilitate and understand protein crystallization. Curr. Opin. Struct. Biol. 2005, 15, 548–555.
- Shim, J.-u.; Cristobal, G.; Link, D.R.; Thorsen, T.; Fraden, S. Using Microfluidics to Decouple Nucleation and Growth of Protein Crystals. Cryst. Growth Des. 2007, 7, 2192–2194.
- Guo, M.T.; Rotem, A.; Heyman, J.A.; Weitz, D.A. Droplet microfluidics for high-throughput biological assays. Lab Chip 2012, 12, 2146–2155.
- Shembekar, N.; Chaipan, C.; Utharala, R.; Merten, C.A. Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics. Lab Chip 2016, 16, 1314–1331.
- Jing, W.; Jiang, X.; Zhao, W.; Liu, S.; Cheng, X.; Sui, G. Microfluidic Platform for Direct Capture and Analysis of Airborne Mycobacterium tuberculosis. Anal. Chem. 2014, 86, 5815–5821.
- Das, D.; Dsouza, A.; Kaur, N.; Soni, S.; Toley, B.J. Paper Stacks for Uniform Rehydration of Dried Reagents in Paper Microfluidic Devices. Sci. Rep. 2019, 9, 15755.
- Luo, J.; Fang, X.; Ye, D.; Li, H.; Chen, H.; Zhang, S.; Kong, J. A real-time microfluidic multiplex electrochemical loop-mediated isothermal amplification chip for differentiating bacteria. Biosens. Bioelectron. 2014, 60, 84–91.
- Mühlig, A.; Bocklitz, T.; Labugger, I.; Dees, S.; Henk, S.; Richter, E.; Andres, S.; Merker, M.; Stöckel, S.; Weber, K.; et al. LOC-SERS: A Promising Closed System for the Identification of Mycobacteria. Anal. Chem. 2016, 88, 7998–8004.
- Lyu, F.; Xu, M.; Cheng, Y.; Xie, J.; Rao, J.; Tang, S.K. Quantitative detection of cells expressing BlaC using droplet-based microfluidics for use in the diagnosis of tuberculosis. Biomicrofluidics 2015, 9, 044120.
- Ip, K.; Chang, J.; Liu, T.; Dou, H.; Lee, G. An integrated microfluidic system for identification of live mycobacterium tuberculosis by real-time polymerase chain reaction. In Proceedings of the 2018 IEEE Micro Electro Mechanical Systems (MEMS), Belfast, UK, 21–25 January 2018; pp. 1124–1127.
- Domínguez, C.M.; Kosaka, P.M.; Sotillo, A.; Mingorance, J.; Tamayo, J.; Calleja, M. Label-Free DNA-Based Detection of Mycobacterium tuberculosis and Rifampicin Resistance through Hydration Induced Stress in Microcantilevers. Anal. Chem. 2015, 87, 1494–1498.
- Zribi, B.; Roy, E.; Pallandre, A.; Chebil, S.; Koubaa, M.; Mejri, N.; Magdinier Gomez, H.; Sola, C.; Korri-Youssoufi, H.; Haghiri-Gosnet, A.M. A microfluidic electrochemical biosensor based on multiwall carbon nanotube/ferrocene for genomic DNA detection of Mycobacterium tuberculosis in clinical isolates. Biomicrofluidics 2016, 10, 014115.
- Islamov, M.; Sypabekova, M.; Kanayeva, D.; Rojas-Solórzano, L. CFD Modeling of Chamber Filling in a Micro-Biosensor for Protein Detection. Biosensors 2017, 7, 45.
- Mbano, I.M.; Mandizvo, T.; Rogich, J.; Kunota, T.T.R.; Mackenzie, J.S.; Pillay, M.; Balagaddé, F.K. Light Forge: A Microfluidic DNA Melting-based Tuberculosis Test. J. Appl. Lab. Med. 2020, 5, 440–453.
- Minero, G.A.S.; Bagnasco, M.; Fock, J.; Tian, B.; Garbarino, F.; Hansen, M.F. Automated on-chip analysis of tuberculosis drug-resistance mutation with integrated DNA ligation and amplification. Anal. Bioanal. Chem. 2020, 412, 2705–2710.
- Law, I.L.G.; Loo, J.F.C.; Kwok, H.C.; Yeung, H.Y.; Leung, C.C.H.; Hui, M.; Wu, S.Y.; Chan, H.S.; Kwan, Y.W.; Ho, H.P.; et al. Automated real-time detection of drug-resistant Mycobacterium tuberculosis on a lab-on-a-disc by Recombinase Polymerase Amplification. Anal. Biochem. 2018, 544, 98–107.
- Li, Y.; Cherukury, H.; Labanieh, L.; Zhao, W.; Kang, D.-K. Rapid Detection of β-Lactamase-Producing Bacteria Using the Integrated Comprehensive Droplet Digital Detection (IC 3D) System. Sensors 2020, 20, 4667.
- Linger, Y.; Knickerbocker, C.; Sipes, D.; Golova, J.; Franke, M.; Calderon, R.; Lecca, L.; Thakore, N.; Holmberg, R.; Qu, P.; et al. Genotyping Multidrug-Resistant Mycobacterium tuberculosis from Primary Sputum and Decontaminated Sediment with an Integrated Microfluidic Amplification Microarray Test. J. Clin. Microbiol. 2018, 56, e01652-17.
- Kukhtin, A.V.; Norville, R.; Bueno, A.; Qu, P.; Parrish, N.; Murray, M.; Chandler, D.P.; Holmberg, R.C.; Cooney, C.G. A Benchtop Automated Sputum-to-Genotype System Using a Lab-on-a-Film Assembly for Detection of Multidrug-Resistant Mycobacterium tuberculosis. Anal. Chem. 2020, 92, 5311–5318.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
15 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




