Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Robert Garry | + 1994 word(s) | 1994 | 2021-09-07 04:24:42 | | | |
| 2 | Rita Xu | -19 word(s) | 1975 | 2021-09-15 04:19:55 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Garry, R. Zika Virus Non-Structural Protein 1. Encyclopedia. Available online: https://encyclopedia.pub/entry/14188 (accessed on 07 February 2026).
Garry R. Zika Virus Non-Structural Protein 1. Encyclopedia. Available at: https://encyclopedia.pub/entry/14188. Accessed February 07, 2026.
Garry, Robert. "Zika Virus Non-Structural Protein 1" Encyclopedia, https://encyclopedia.pub/entry/14188 (accessed February 07, 2026).
Garry, R. (2021, September 14). Zika Virus Non-Structural Protein 1. In Encyclopedia. https://encyclopedia.pub/entry/14188
Garry, Robert. "Zika Virus Non-Structural Protein 1." Encyclopedia. Web. 14 September, 2021.
Copy Citation
Infection with Zika virus (ZIKV), a member of the Flavivirus genus of the Flaviviridae family, typically results in mild self-limited illness, but severe neurological disease occurs in a limited subset of patients.
Zika virus
non-structural protein 1
site-directed mutagenesis
polyclonal antibodies
1. Introduction
Zika virus (ZIKV) is named after the Zika forest in Uganda, where it was first isolated from a sentinel monkey in 1947 [1], and shortly after from mosquitoes in the same area [2]. Follow-up serological studies revealed Zika to be widespread in Africa and Asia [1][3][4]. The first reported natural ZIKV infection in humans was reported in 1964, when a scientist was infected while isolating virus from mosquitos in Uganda [4][5]. The first known incidence of Zika outside Africa and Asia was in 2007 in Yap State, Micronesia [6], in what became the first large outbreak recorded [7]. A total of 73% of residents of Yap Island 3 years old or older were infected [8]. In 2013, cases of Zika began being reported in French Polynesia. A total of 8746 cases were estimated to have occurred during this outbreak [9]. Other ZIKV outbreaks included New Caledonia, Cook Islands and Easter Island [10].
Beginning in early 2015, cases of ZIKV infection were reported in Bahia, Brazil [11]. A second introduction of Zika into Brazil occurred at about the same time, in Natal [12]. Estimates were from 440,000 to 1.3 million cases [13], with an eventual estimate of over 500,000 cases of Zika [14] and 73% of the population exposed in some areas [15]. Other countries in the Americas also reported Zika cases during this time. Colombia reported over 100,000 suspected cases [16], with a total of 27 countries in the Americas reporting cases, including Mexico, Guatemala, the Dominican Republic and Panama [17]. Eventually, Zika cases were reported in the continental United States, with multiple introductions of ZIKV into Florida [18], as well as Texas. These cases in Texas and Florida were locally acquired, indicating at least some level of circulation in local mosquito populations [19]. Afterward, ZIKV made its way to Cape Verde, Samoa, the Solomon Islands and the surrounding region [5][20]. In all, 87 countries were affected by the pandemic, with cases continuing after the large outbreak, including 30,000 cases in 2018 [14]. ZIKV has established itself as endemic to many more regions than it was prior to the outbreak and may continue to infect humans routinely into the future.
ZIKV infection usually results in a self-limited illness [21]. In symptomatic cases the most common symptom is a maculopapular rash, occurring in over 80% of cases [22], often on the face, torso and upper arms [23][24]. Another common symptom is arthralgia in about 62% of cases, mainly involving small joints [22][25], and occasionally larger joints as well [24][26]. Conjunctivitis is seen in roughly half of patients, with both eyes commonly being affected [22][27]. Other symptoms may be seen, such as myalgia, headache, nausea, vomiting, sore throat and cough [28]. If present, symptoms resolve within a week and are generally not fatal [29], provided there is not a comorbidity, such as sickle cell anemia [30]. Before 2013, no significant complications were reported from infections with ZIKV [4]. However, it was later determined that ZIKV is capable of establishing persistent infections, crossing placental and neuronal barriers, and damaging neurons [31]. During the French Polynesia outbreak, multiple cases of Guillain–Barré Syndrome (GBS) were reported [5][32][33]. GBS is a neurological disorder presenting as a rapid ascending paralysis that results in respiratory failure [34]. Another complication seen from ZIKV infections, microcephaly, occurs in a developing fetus in infected pregnant women. Defined as a neurological malformation resulting in decreased head circumference due to death of neural progenitor cells, microcephaly has been implicated as a result of ZIKV infections in eight individuals during the French Polynesia outbreak [35]. The outbreak in Brazil resulted in many more reports of congenital microcephaly, prompting the World Health Organization to declare a Public Health Emergency of International Concern on 1 February 2016 [36]. Other complications include conditions such as optic neuropathy, uveitis and congenital glaucoma resulting in loss of vision [37][38].
Antigen-capture diagnostic assays targeting NS1 have been utilized in the past for dengue virus (DENV) infection, with NS1 being established as an early biomarker for flavivirus infection [39][40][41][42]. The flavivirus non-structural protein 1 (NS1) functions in genome replication as an intracellular dimer and in immune system evasion as a secreted hexamer. Assays are often in Enzyme Linked Immunosorbent Assay (ELISA) format using monoclonal antibodies (mAbs) to overcome the genetic similarity among flaviviruses [43][44][45][46][47]. One of the assays commonly employed for the early diagnosis of dengue is the InBios DENV Detect NS1 ELISA Kit. It has a reported sensitivity of 86.8% and specificity of 97.8% [48]. One potential issue may be the lack of sensitivity in secondary dengue cases, with one study indicating 100% sensitivity in primary infections, and 10% sensitivity in secondary infections [41]. This is likely due to circulating anti-NS1 antibodies present during secondary infections [49]. This demonstrated success with NS1-based detection of DENV infection offers an attractive option for detection of infection with other flaviviruses.
2. Computational Analysis of Flavivirus NS1
Crystal structures for full-length, glycosylated NS1 from WNV and DENV have been reported (PDB: 5GS6, 5IY3). These structures define protein domains and potential immunoreactive sites (Figure 1). NS1 is a dimer with three major domains in each monomer (Figure 1A). Due to the sequence similarity between NS1 proteins of flaviviruses, the ZIKV NS1 is expected to be structurally similar to the NS1 proteins of WNV, DENV and other members of the flavivirus genus of the Flaviviridae. A small “β-roll” dimerization domain (amino acids 1–29, red) links the two monomers of the NS1 dimer. The wing domain (amino acids 30–180, yellow) of each monomer extends from the central β-roll. A discontinuous connector subdomain (amino acids 30–37 and 152–180, orange) is also linked to the dimerization domain. A major feature of NS1 is the third domain (amino acids 181–352, blue), with a ladder-like feature extending along the length of the dimer and comprised of 18 β-strands. Most of the inter-strand loops are short with the notable exception of a long “loop” (green) between amino acids 219–272 that lacks secondary structure but has extensive hydrogen bonding.
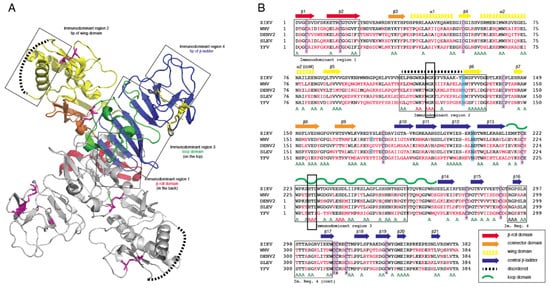
Figure 1. Comparison of non-structural protein 1 sequences among closely related flaviviruses. (A): Ribbon model highlighting regions of the NS1 protein containing segments exposed at the outer surface to the host environment. (B): Sequence comparison showing regions with high sequence disparity. Amino acids depicted in red differ from the corresponding ZIKV NS1 amino acids. A represents positions with two sequences with amino acids identical to ZIKV NS1. The boxes highlight highly conserved sequences, amino acids 117–119 and 227–229, that were mutated to alanine in immunodominant regions 2 and 3.
Previous studies have identified several immunodominent regions of the protein (Figure 1A,B). Immunodominent region 1 maps to the β-roll, immunodominant region 2 to a disordered sequence at the tip of the wing domain, immunodominent region 3 to the loop and immunodominant region 4 to the ends of the β-sheet ladder. Of these immuno-dominent regions 2 and 3 display the most sequence variability between flavivirus NS1 proteins. Site-directed mutagenesis was performed to generate mutated rNS1. Primers were designed to mutate residues in regions of NS1 with high sequence similarity to other members of the Flaviviridae family. Mutations were targeted toward sequences in the protein in immunodominant regions 2 and 3 that shared sequence identity with related flaviviruses. These regions were selected because they are the most highly conserved sequences in any of the immunodominant regions. Mutations of the sequence were introduced to the ZIKV NS1 sequence as triple alanine repeats to minimize impacts on folding of the protein structure. Specific mutations (117–119 and 227–229) were designed for lower reactivity of the protein to antibodies to non-ZIKV Flavivirus NS1 proteins and increase specificity of a diagnostic toward ZIKV.
3. Production of Mutant Zika Virus NS1 and Dengue Virus NS1
To produce recombinant non-structural protein 1 (rNS1) for the variant NS1, as well as the related DENV, sequences were amplified and cloned into the pET-45b(+) vector. The pET-45b(+) vector enables addition of an N-terminal His6 tag that was used for purification (Figure 2). After induction of protein production, multiple colonies were screened for expression by SDS-PAGE, with expression appearing to be near peak at 4 h post induction. Upon expression, localization was confirmed by sonication of an induced pellet followed by centrifugation to pellet insoluble mass and resuspension of that mass. This revealed the vast majority of protein to be in the insoluble fraction (inclusion body). Solubilization of the inclusion body was performed using urea-based buffers in order to denature the protein and pull it into solution. Following the protein through multiple purification steps shows the protein is not soluble in either the initial Triton X-100 solubilization buffer, or 1 M urea washes, but must be solubilized in 8 M urea. Ni-NTA purification of protein based on the added hexahistidine tag was performed on the 8 M urea soluble fraction with washes using 20 mM imidazole to eliminate non-specific binding. Elution was performed with 250 mM imidazole to outcompete the binding of the protein to the resin and allow the protein to flow through the column.
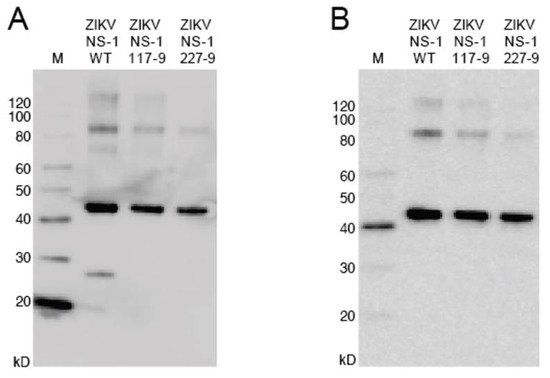
Figure 2. Western blot of NS1 mutants. (A): The blot was probed with anti-His6 antibody targeted toward the protein N-terminus. (B): The blot was probed with anti-ZIKV NS1 monoclonal antibody targeted toward the C-terminus.
4. Reactivity of the rNS1 to Antibodies Circulating in Patient Serum and Immunized Goats
To compare the seroreactivity of the wild-type rNS1 to the mutant rNS1 samples from Zika patients from Columbia and the Dominican Republic were tested (Figure 3). IgG binding ratios for the Colombia samples show no differences between wild-type NS1 and either mutant protein. IgM binding ratios for the samples from the Columbian patients indicate increased binding to the wild-type protein. Samples from the Dominican Republic patients showed a different trend, with increased binding to the ZIKV NS1 117–119 and decreased binding to the ZIKV NS1 227–229. Both mutant NS1 proteins bound serum IgM more readily in the Dominican Republic samples compared to wild-type NS1 (Figure 3). Differences in protein–protein interactions or other structural changes in the mutant proteins could account for the differential antibody binding compared to wild-type NS1.
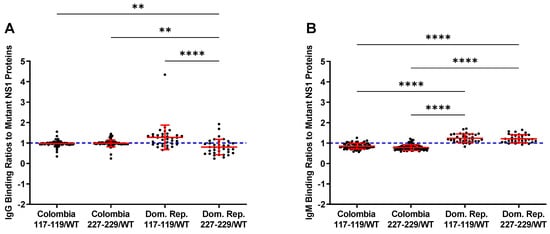
Figure 3. Ratios of mutant vs. wild-type binding of NS1 by patient samples. The blue line at 1 indicates equal binding. Numbers over 1 indicate increased binding to the named mutant, while under 1 indicate increased wild-type binding. (A): Ratios of mutant/wild-type binding from IgG in samples show preferential binding to 117–119 mutant NS1. (B): IgM shows preferential binding of mutant NS1 proteins in samples from the Dominican Republic. Colombia = Colombia samples from suspected ZIKV infection. Dom Rep = Dominican Republic samples from suspected ZIKV infection. 117–119 = ZIKV NS1 W117A, G118A, K119A. 227–229 = ZIKV NS1 H227A, T228A, L229A Comparisons were made using Kruskal–Wallis ANOVA. Asterisks represent significant comparisons (**** p < 0.0001; ** p < 0.01).
As a large volume of goat serum would be required to produce antigen-capture ELISA in sufficient amounts for the current and future studies sera from two goats immunized with either wild-type or mutant rNS1 were pooled. Differences in reactivity are present significantly in the pooled serum from the immunized goats. Goats immunized with the wild-type protein produce antibodies that bound to all three variants of ZIKV NS1, as well as DENV NS1, less strongly than antiserum produced from either mutant ZIKV NS1 proteins (Figure 4). The reduced reactivity, along with the data indicating high binding of the wild-type protein with antiserum from mutants, reinforces the idea that there are significant differences in the protein–protein interactions or structural differences of the mutate rNS1 compared to the wild-type protein. The mutant rNS1 bound each of the other proteins in amounts not significantly different, indicating that the mutations had little effect on binding of NS1, at least in goats.
Figure 4. Serum from goats immunized with ZIKV mutants show differential binding to NS1 proteins. Antibody-capture ELISA was performed on a mixture of serum from two goats immunized with ZIKV NS1 protein. Binding of WT ZIKV NS1 showed lower binding to WT (A), both mutants (B,C), as well as DENV2 WT NS1 (D). Data represent samples run in duplicate. Error bars (standard error of the mean) were smaller than the symbols as drawn. The experiment was repeated a total of three times. The legend in panel (D) also applies to panels (A–C).
References
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520.
- Dick, G.W.A.; Haddow, A.J. Uganda S virus: A hitherto unrecorded virus isolated from mosquitoes in Uganda. I. Isolation and pathogenicity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 600–618.
- Smithburen, K.C.; Kerr, J.A.; Gatne, P.B. Neutralizing antibodies against certain viruses in the sera of residents of India. J. Immunol. 1954, 72, 248–257.
- Simpson, D.I.H. Zika virus infection in man. Trans. R. Soc. Trop. Med. Hyg. 1964, 58, 335–338.
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 2016, 16, e119–e126.
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239.
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.M. Rapid spread of emerging Zika virus in the Pacific area. Clin. Microbiol. Infect. 2014, 20, O595–O596.
- Duffy, M.R.; Chen, T.-H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543.
- Roth, A.; Mercier, A.; Lepers, C.; Hoy, D.; Duituturaga, S.; Benyon, E.; Guillaumot, L.; Souares, Y. Concurrent outbreaks of dengue, chikungunya and Zika virus infections—An unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012–2014. Euro Surveill. 2014, 19, 1–8.
- Tognarelli, J.; Ulloa, S.; Villagra, E.; Lagos, J.; Aguayo, C.; Fasce, R.; Parra, B.; Mora, J.; Becerra, N.; Lagos, N.; et al. A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch. Virol. 2016, 161, 665–668.
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg. Infect. Dis. 2015, 21, 1885–1886.
- Zanluca, C.; de Melo, V.C.A.; Mosimann, A.L.P.; Dos Santos, G.I.V.; Dos Santos, C.N.D.; Luz, K. First report of autochthonous transmission of Zika virus in Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 569–572.
- Bogoch, I.I.; Brady, O.J.; Kraemer, M.U.G.; German, M.; Creatore, M.I.; Kulkarni, M.A.; Brownstein, J.S.; Mekaru, S.R.; Hay, S.I.; Groot, E.; et al. Anticipating the international spread of Zika virus from Brazil. Lancet 2016, 387, 335–336.
- Musso, D.; Ko, A.I.; Baud, D. Zika Virus Infection—After the Pandemic. N. Engl. J. Med. 2019, 381, 1444–1457.
- Rodriguez-Barraquer, I.; Costa, F.; Nascimento, E.J.M.; Nery, N.; Castanha, P.M.S.; Sacramento, G.A.; Cruz, J.; Carvalho, M.; De Olivera, D.; Hagan, J.E.; et al. Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science 2019, 363, 607–610.
- Arrieta, G.; Mattar, S.; Villero-Wolf, Y.; Gomezcaceres, L.; Doria, A. Evaluation of serological test of Zika in an endemic area of flavivirus in the Colombian Caribbean. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 29.
- Paixão, E.S.; Barreto, F.; Da Glória Teixeira, M.; Da Conceição N Costa, M.; Rodrigues, L.C. History, epidemiology, and clinical manifestations of Zika: A systematic review. Am. J. Public Health 2016, 106, 606–612.
- Grubaugh, N.D.; Ladner, J.T.; Kraemer, M.U.G.; Dudas, G.; Tan, A.L.; Gangavarapu, K.; Wiley, M.R.; White, S.; Thézé, J.; Magnani, D.M.; et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 2017, 546, 401–405.
- Adams, L.E.; Martin, S.W.; Lindsey, N.P.; Lehman, J.A.; Rivera, A.; Kolsin, J.; Landry, K.; Staples, J.E.; Sharp, T.M.; Paz-Bailey, G.; et al. Epidemiology of Dengue, Chikungunya, and Zika Virus Disease in U.S. States and Territories, 2017. Am. J. Trop. Med. Hyg. 2019, 101, 884–890.
- Basu, R.; Tumban, E. Zika virus on a spreading spree: What we now know that was unknown in the 1950’s. Virol. J. 2016, 13, 165.
- Moghadas, S.M.; Shoukat, A.; Espindola, A.L.; Pereira, R.S.; Abdirizak, F.; Laskowski, M.; Viboud, C.; Chowell, G. Asymptomatic Transmission and the Dynamics of Zika Infection. Sci. Rep. 2017, 7, 5829.
- Salehuddin, A.R.; Haslan, H.; Mamikutty, N.; Zaidun, N.H.; Azmi, M.F.; Senin, M.M.; Syed Ahmad Fuad, S.B.; Thent, Z.C. Zika virus infection and its emerging trends in Southeast Asia. Asian Pac. J. Trop. Med. 2017, 10, 211–219.
- Hayes, E.B. Zika virus outside Africa. Emerg. Infect. Dis. 2009, 15, 1347–1350.
- Foy, B.D.; Kobylinski, K.C.; Chilson Foy, J.L.; Blitvich, B.J.; Travassos da Rosa, A.; Haddow, A.D.; Lanciotti, R.S.; Tesh, R.B. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg. Infect. Dis. 2011, 17, 880–882.
- Tappe, D.; Rissland, J.; Gabriel, M.; Emmerich, P.; Gunther, S.; Held, G.; Smola, S.; Schmidt-Chanasit, J. First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Euro Surveill. 2014, 19, 1–4.
- Ginier, M.; Neumayr, A.; Günther, S.; Schmidt-Chanasit, J.; Blum, J. Zika without symptoms in returning travellers: What are the implications? Travel Med. Infect. Dis. 2016, 14, 16–20.
- Goorhuis, A.; von Eije, K.J.; Douma, R.A.; Rijnberg, N.; van Vugt, M.; Stijnis, C.; Grobusch, M.P. Zika virus and the risk of imported infection in returned travelers: Implications for clinical care. Travel Med. Infect. Dis. 2016, 14, 13–15.
- Colombo, T.E.; Estofolete, C.F.; Reis, A.F.N.; da Silva, N.S.; Aguiar, M.L.; Cabrera, E.M.S.; Dos Santos, I.N.P.; Costa, F.R.; Cruz, L.E.A.A.; Rombola, P.L.; et al. Clinical, laboratory and virological data from suspected ZIKV patients in an endemic arbovirus area. J. Clin. Virol. 2017, 96, 20–25.
- Karkhah, A.; Nouri, H.R.; Javanian, M.; Koppolu, V.; Masrour-roudsari, J. Zika virus: Epidemiology, clinical aspects, diagnosis, and control of infection. Eur. J. Clin. Microbiol. Infect Dis. 2018, 37, 2035–2043.
- Arzuza-Ortega, L.; Polo, A.; Pérez-Tatis, G.; López-García, H.; Parra, E.; Pardo-Herrera, L.C.; Rico-Turca, A.M.; Villamil-Gómez, W.; Rodríguez-Morales, A.J. Fatal Sickle Cell Disease and Zika Virus Infection in Girl from Colombia. Emerg. Infect. Dis. 2016, 22, 925–927.
- Mladinich, M.C.; Schwedes, J.; Mackow, E.R. Zika Virus Persistently Infects and Is Basolaterally Released from Primary Human Brain Microvascular Endothelial Cells. mBio 2017, 8, e00952-17.
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastère, S.; Valour, F.; Baudouin, L.; Mallet, H.P.; Musso, D.; Ghawche, F. Zika virus infection complicated by Guillain-Barré syndrome—Case report, French Polynesia, December 2013. Euro Surveill. 2014, 19, 7–9.
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teisiier, A.; Larre, P.; et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet 2016, 387, 1531–1539.
- Willison, H.J.; Jacobs, B.C.; van Doorn, P.A. Guillain-Barré syndrome. Lancet 2016, 388, 717–727.
- Cauchemez, S.; Besnard, M.; Bompard, P.; Dub, T.; Guillemette-Artur, P.; Eyrolle-Guignot, D.; Salje, H.; Van Kerkhove, M.D.; Abadie, V.; Garel, C.; et al. Association between Zika virus and microcephaly in French Polynesia, 2013–2015: A retrospective study. Lancet 2016, 387, 2125–2132.
- WHO Director-General Summarizes the Outcome of the Emergency Committee Regarding Clusters of Microcephaly and Guillain-Barré Syndrome. Available online: https://www.who.int/en/news-room/detail/01-02-2016-who-director-general-summarizes-the-outcome-of-the-emergency-committee-regarding-clusters-of-microcephaly-and-guillain-barré-syndrome (accessed on 1 December 2020).
- De Moraes, C.G.; Pettito, M.; Yepez, J.B.; Sakuntabhai, A.; Simon-Loriere, E.; Zaidi, M.B.; Prot, M.; Ruffie, C.; Kim, S.S.; Allikmets, R.; et al. Optic neuropathy and congenital glaucoma associated with probable Zika virus infection in Venezuelan patients. JMM Case Rep. 2018, 5, e005145.
- Manangeeswaran, M.; Kielczewski, J.L.; Sen, H.N.; Xu, B.C.; Ireland, D.D.C.; McWilliams, I.L.; Chan, C.-C.; Caspi, R.R.; Verthelyi, D. ZIKA virus infection causes persistent chorioretinal lesions. Emerg. Microbes Infect. 2018, 7, 96.
- Allonso, D.; Meneses, M.D.F.; Fernandes, C.A.; Ferreira, D.F.; Mohana-Borges, R. Assessing positivity and circulating levels of NS1 in samples from a 2012 dengue outbreak in Rio de Janeiro, Brazil. PLoS ONE 2014, 9, 6–8.
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131.
- Young, P.R.; Hilditch, P.A.; Bletchly, C.; Halloran, W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 2000, 38, 1053–1057.
- Wang, S.M.; Sekaran, S.D. Early diagnosis of dengue infection using a commercial Dengue Duo Rapid Test Kit for the detection of NS1, IgM, and IgG. Am. J. Trop. Med. Hyg. 2010, 83, 690–695.
- Hu, D.; Fry, S.R.; Huang, J.X.; Ding, X.; Qiu, L.; Pan, Y.; Chen, Y.; Jin, J.; McElnea, C.; Buechler, J.; et al. Comparison of surface plasmon resonance, resonant waveguide grating biosensing and enzyme linked immunosorbent assay (ELISA) in the evaluation of a dengue virus immunoassay. Biosensors 2013, 3, 297–311.
- Sinawang, P.D.; Rai, V.; Ionescu, R.E.; Marks, R.S. Electrochemical lateral flow immunosensor for detection and quantification of dengue NS1 protein. Biosens. Bioelectron. 2016, 77, 400–408.
- Linares, E.M.; Pannuti, C.S.; Kubota, L.T.; Thalhammer, S. Immunospot assay based on fluorescent nanoparticles for Dengue fever detection. Biosens. Bioelectron. 2013, 41, 180–185.
- Antunes, P.; Watterson, D.; Parmvi, M.; Burger, R.; Boisen, A.; Young, P.; Cooper, M.A.; Hansen, M.F.; Ranzoni, A.; Donolato, M. Quantification of NS1 dengue biomarker in serum via optomagnetic nanocluster detection. Sci. Rep. 2015, 5, 1–10.
- Lee, J.; Kim, H.; Chong, C.; Song, H. Development and clinical evaluation of a highly accurate dengue NS1 rapid test: From the preparation of a soluble NS1 antigen to the construction of an RDT. Diagn. Microbiol. Infect. Dis. 2015, 82, 128–134.
- InBios DENV Detect NS1 ELISA Product Page. Available online: https://inbios.https//inbios.com/denv-detect-ns1-elisa-usa/com/denv-detect-ns1-elisa-usa/ (accessed on 11 December 2019).
- Shu, P.; Huang, J. Current advances in dengue diagnosis. Clin. Diagn. Lab. Immunol. 2004, 11, 642–650.
More
Information
Subjects:
Virology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
858
Revisions:
2 times
(View History)
Update Date:
15 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




