
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mourad A. M. Aboul-Soud | + 4102 word(s) | 4102 | 2021-09-07 10:18:32 |
Video Upload Options
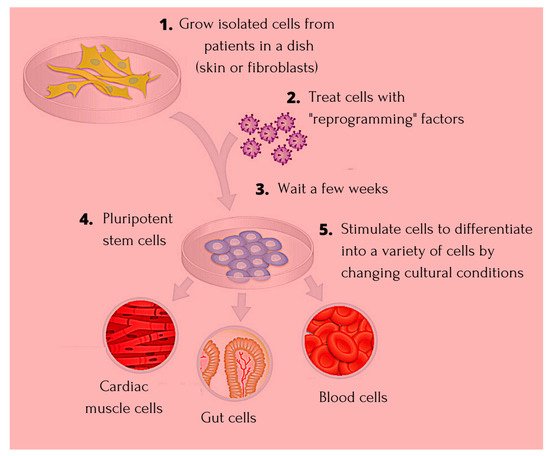
The discovery of induced pluripotent stem cells (iPSCs) has made an invaluable contribution to the field of regenerative medicine, paving way for identifying the true potential of human embryonic stem cells (ESCs). iPSCs have been widely used in cardiac disease modelling, studying inherited arrhythmias, neural disorders including Alzheimer’s disease, liver disease, and spinal cord injury. Extensive research around identifying factors that are involved in maintaining the identity of ESCs during induction of pluripotency in somatic cells is undertaken.
1. Introduction
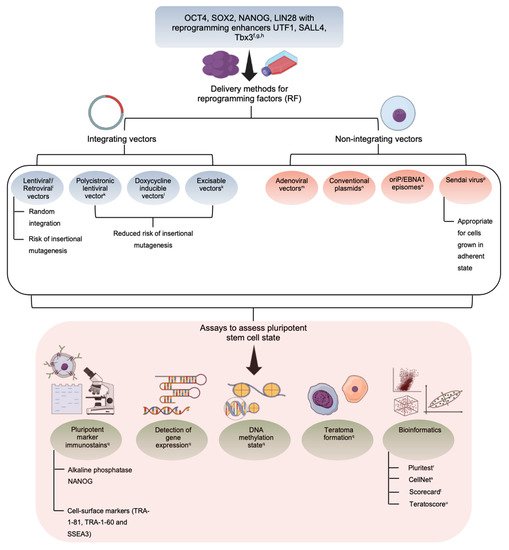
|
Vector |
Cell Type |
Genes |
Efficiency |
Reference |
|---|---|---|---|---|
|
Retrovirus |
Skin fibroblasts |
OCT4, SOX2, KLF4 |
0.001% |
[18] |
|
Fibroblasts |
OCT4, SOX2 and Valproic acid |
0.001% |
[19] |
|
|
Skin cancer cell line |
miR-302 |
Unknown |
[20] |
|
|
Lentivirus |
Embryonic fibroblasts |
OCT4, SOX2, NANOG, LIN28 |
0.01% |
[21] |
|
Fibroblasts |
OCT4, SOX2, KLF4, c-MYC |
0.01% |
[22] |
|
|
Adenovirus |
Embryonic fibroblasts |
OCT4, SOX2, KLF4, c-MYC |
0.0002% |
[23] |
|
Sendai virus |
Cord blood CD34+ cells |
OCT4, SOX2, KLF4, c-MYC |
0.2% |
[24] |
|
Recombinant protein |
Fibroblasts |
OCT4, SOX2, KLF4, c-MYC |
0.001% |
[25] |
|
mRNA |
Fibroblasts |
OCT4, SOX2, NANOG, LIN28 |
0.05% |
[26] |
2. Induced Pluripotent Stem Cells—The Niche Favoring Unique Aspects
3. Application of iPSC in Cardiac Disease
4. Application of iPSC in Degenerative Diseases
5. Application of iPSC in Blood Disorders
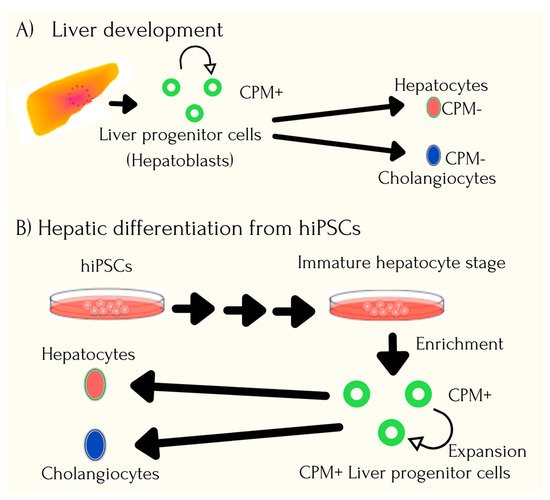
6. Application of iPSC in Organ Dysfunctions
7. Application of iPSCs in Cancer Syndromes
References
- Weismann, A. The Germ-Plasm: A Theory of Heredity; Charles Scribner’s Sons: New York, NY, USA, 1893.
- Gurdon, J.B.; Byrne, J.A. The first half-century of nuclear transplantation. Proc. Natl. Acad. Sci. USA 2003, 100, 8048–8052.
- Bradley, A.; Evans, M.; Kaufman, M.H.; Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 1984, 309, 255–256.
- Cowan, C.A.; Atienza, J.; Melton, D.A.; Eggan, K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 2005, 309, 1369–1373.
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced pluripotent stem cells and their use in human models of disease and development. Physiol. Rev. 2018, 99, 79–114.
- Abbar, A.A.; Ngai, S.C.; Nograles, N.; Alhaji, S.Y.; Abdullah, S. Induced pluripotent stem cells: Reprogramming platforms and applications in cell replacement therapy. BioRes. Open Access 2020, 9, 121–136.
- Nishikawa, S.; Goldstein, R.A.; Nierras, C.R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. Mol. Cell Biol. 2008, 9, 725–729.
- Ye, L.; Ni, X.; Zhao, Z.-A.; Lei, W.; Hu, S. The application of induced pluripotent stem cells in cardiac disease modelling and drug testing. J. Cardiovasc. Transl. Res. 2018, 11, 366–374.
- Mandai, M.; Watanabe, A.; Kurimoto, Y.; Hirami, Y.; Morinaga, C.; Daimon, T.; Fujihara, M.; Akimaru, H.; Sakai, N.; Shibata, Y.; et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017, 376, 1038–1046.
- Sundberg, M.; Bogetofte, H.; Lawson, T.; Jansson, J.; Smith, G.; Astradsson, A.; Moore, M.; Osborn, T.; Cooper, O.; Spealman, R.; et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells 2013, 31, 1548–1562.
- Sridharan, R.; Tchieu, J.; Mason, M.J.; Yachechko, R.; Kuoy, E.; Horvath, S.; Zhou, Q.; Plath, K. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009, 136, 364–377.
- Winkler, T.; Cantilena, A.; Metais, J.-Y.; Xu, X.; Nguyen, A.-D.; Borate, B.; Antosiewicz-Bourget, J.E.; Wolfsberg, T.G.; Thomson, J.A.; Dunbar, C.E. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells 2010, 28, 687–694.
- Somers, A.; Jean, J.-C.; Sommer, C.A.; Omari, A.; Ford, C.C.; Mills, J.A.; Ying, L.; Sommer, A.G.; Jean, J.M.; Smith, B.W.; et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 2010, 28, 1728–1740.
- Stadtfeld, M.; Hochedlinger, K. Induced pluripotency: History, mechanisms, and applications. Genes Dev. 2010, 24, 2239–2263.
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953.
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zeng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017, 4, 43–63.
- Ong, S.-G.; Lee, W.H.; Kodo, K.; Wu, J.C. MicroRNA-mediated regulation of differentiation and trans-differentiation in stem cells. Adv. Drug Deliv. Rev. 2015, 88, 3–15.
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106.
- Huangfu, D.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275.
- Lin, S.-L.; Chang, D.C.; Chang-Lin, S.; Lin, C.-H.; Wu, D.T.S.; Chen, D.T.; Ying, S.-Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124.
- Yu, Y.; Wang, X.; Nyberg, S.L. Application of induced pluripotent stem cells in liver diseases. Cell Med. 2014, 7, 1–13.
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872.
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 2667–2674.
- Nishishita, N.; Takenaka, C.; Fusaki, N.; Kawamata, S. Generation of human induced pluripotent stem cells from cord blood cells. J. Stem Cells 2011, 6, 101–108.
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming factors. Cell Stem Cell 2009, 4, 472–476.
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193.
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63.
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.-K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77.
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog is the gateway to the pluripotent ground state. Cell 2009, 138, 722–737.
- Gidekel, S.; Bergman, Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002, 277, 34521–34530.
- Hanna, J.; Wernig, M.; Markoulaki, S.; Sun, C.-W.; Meissner, A.; Cassady, J.P.; Beard, C.; Brambrink, T.; Wu, L.-C.; Townes, T.M.; et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 2007, 318, 1920–1923.
- Doi, D.; Magotani, H.; Kikuchi, T.; Ikeda, M.; Hiramatsu, S.; Yoshida, K.; Amano, N.; Nomura, M.; Umekage, M.; Morizane, A.; et al. Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 2020, 11, 3369.
- Nenasheva, T.; Gerasimova, T.; Serdyuk, Y.; Grigoreva, E.; Kosmiadi, G.; Nikolaev, A.; Dashinimaev, E.; Lyadova, I. Macrophages derived from human induced pluripotent stem cells are low-activated “naïve-like” cells capable of restricting Mycobacteria growth. Front. Immunol. 2020, 11, 1016.
- Nagashima, T.; Shimizu, K.; Matsumoto, R.; Honda, H. Selective elimination of human induced pluripotent stem cells using medium with high concentration of L-alanine. Sci. Rep. 2018, 8, 12427.
- Matsumoto, R.; Shimizu, K.; Nagashima, T.; Tanaka, H.; Mizuno, M.; Kikkawa, F.; Hori, M.; Honda, H. Plasma-activated medium selectively eliminates undifferentiated human induced pluripotent stem cells. Regen. Ther. 2016, 5, 55–63.
- Burkert, K.; Taheri, H.; Hamad, S.; Oliverio, M.; Peinkofer, G.; Kornfeld, J.-W.; Harnying, W.; Pfannkuche, K.; Hescheler, J.; Berkessel, A.; et al. Salicylic diamines selectively eliminate residual undifferentiated cells from pluripotent stem cell-derived cardiomyocyte preparations. Sci. Rep. 2021, 11, 2391.
- Wu, J.C.; Garg, P.; Yoshida, Y.; Yamanaka, S.; Gepstein, L.; Hulot, J.-S.; Knollmann, B.C.; Schwartz, P.J. Towards precision medicine with human iPSCs for cardiac channelopathies. Circ. Res. 2019, 125, 653–658.
- Kumar, S.; Blangero, J.; Curran, J.E. Induced pluripotent stem cells in disease modelling and gene identification. Methods Mol. Biol. 2018, 1706, 17–38.
- Wen, Z.; Nguyen, H.N.; Guo, Z.; Lalli, M.A.; Wang, X.; Su, Y.; Kim, N.-S.; Yoon, K.-J.; Shin, J.; Zhang, C.; et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014, 515, 414–418.
- Wong, I.Y.; Poon, M.-W.; Pang, R.T.; Lian, Q.; Wong, D. Promises of stem cell therapy for retinal degenerative diseases. Graefe’s Arch. Clin. Exp. Ophthalmol. 2011, 249, 1439–1448.
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated inti motor neurons. Science 2008, 321, 1218–1221.
- Raya, A.; Rodriguez-Piza, I.; Guenechea, G.; Vassena, R.; Navarro, S.; Barrero, M.J.; Consiglio, A.; Castella, M.; Rio, P.; Sleep, E.; et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 2009, 460, 53–59.
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977.
- D’Amour, K.A.; Bang, A.G.; Eliazer, S.; Kelly, O.G.; Agulnick, A.D.; Smart, N.G.; Moorman, M.A.; Kroon, E.; Carpenter, M.K.; Baetge, E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006, 24, 1392–1401.
- Kondo, Y.; Toyoda, T.; Inagaki, N.; Osafune, K. iPSC technology-based regenerative therapy for diabetes. J. Diabetes Investig. 2018, 9, 234–243.
- Sahu, S.; Hemlata; Verma, A. Adverse events related to blood transfusion. Indian J. Anaesth. 2014, 58, 543–551.
- Seo, Y.; Shin, K.-H.; Kim, H.H.; Kim, H.-S. Current advances in red blood cell generation using stem cells from diverse sources. Stem Cells Int. 2019, 2019, 9281329.
- Chang, K.-H.; Bonig, H.; Papayannopoulou, T. Generation and characterization of erythroid cells from human embryonic stem cells and induced pluripotent stem cells: An overview. Stem Cells Int. 2011, 2011, 791604.
- Ebrahimi, M.; Forouzesh, M.; Raoufi, S.; Ramazii, M.; Ghaedrahmati, F.; Farzaneh, M. Differentiation of human induced pluripotent stem cells into erythroid cells. Stem Cell Res. Ther. 2020, 11, 483.
- Peyrard, T.; Bardiaux, L.; Krause, C.; Kobari, L.; Lapillonne, H.; Andreu, G.; Douay, L. Banking of pluripotent adult stem cells as an unlimited source for red blood cell production: Potential applications for alloimmunized patients and rare blood challenges. Transfus. Med. Rev. 2011, 25, 206–216.
- Ding, Q.; Cowan, C.A. Liver in a dish. Cell Res. 2013, 23, 1242–1243.
- Fox, I.J.; Duncan, S.A. Engineering liver tissue from induced pluripotent stem cells: A first step in generating new organs for transplantation? Hepatology 2013, 58, 2198–2201.
- Willenbring H and Soto-Gutierrez, A. Transplantable liver organoids made from only three ingredients. Cell Stem Cell 2013, 13, 139–140.
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.-R.; Ueno, Y.; Zheng, Y.-W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484.
- Chang, C.-J.; Bouhassira, E.E. Zinc-finger nuclease-mediated correction of alpha-thalassemia in iPS cells. Blood 2012, 120, 3906–3914.
- Zhou, R.; Xu, A.; Tu, J.; Liu, M.; Gingold, J.A.; Zhao, R.; Lee, D.F. Modeling osteosarcoma using Li-Fraumeni syndrome patient-derived induced pluripotent stem cells. JOVE 2018, 136, 57664.
- Mulero-Navarro, S.; Sevilla, A.; Roman, A.C.; Lee, D.-F.; D’Souza, S.L.; Pardo, S.; Riess, I.; Su, J.; Cohen, N.; Schaniel, C.; et al. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11–associated juvenile myelomonocytic leukemia. Cell Rep. 2015, 13, 504–515.
- Portier, L.; Desterke, C.; Chaker, D.; Oudrhiri, N.; Asgarova, A.; Dkhissi, F.; Turhan, A.G.; Bennaceur-Griscelli, A.; Griscelli, F. iPSC-derived hereditary breast cancer model reveals the BRCA1-deleted tumor niche as a new culprit in disease progression. Int. J. Mol. Sci. 2021, 22, 1227.
- Chao, H.-M.; Chern, E. Patient-derived induced pluripotent stem cells for models of cancer and cancer stem cell research. J. Formos. Med. Assoc. 2018, 117, 1046–1057.
- Reyal, F.; Guyader, C.; Decraene, C.; Lucchesi, C.; Auger, N.; Assayag, F.; De Plater, L.; Gentien, D.; Poupon, M.-F.; Cottu, P.; et al. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. 2012, 14, R11.
- Krumbach, R.; Schuler, J.; Hofmann, M.; Giesemann, T.; Fiebig, H.-H.; Beckers, T. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: Activation of MET as one mechanism for drug resistance. Eur. J. Cancer 2011, 47, 1231–1243.
- Chen, Z.; Trotman, L.C.; Shaffer, D.; Lin, H.-K.; Dotan, Z.A.; Niki, M.; Koutcher, J.A.; Scher, H.I.; Ludwig, T.; Gerald, W.; et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005, 436, 725–730.
- Wang, L.; Pegram, M.D.; Wu, J.C. Induced pluripotent stem cells as a novel cancer vaccine. Expert Opin. Biol. Ther. 2019, 19, 1191–1197.




