Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gerardo Cazzato | + 1105 word(s) | 1105 | 2021-09-06 05:57:58 | | | |
| 2 | Camila Xu | + 100 word(s) | 1205 | 2021-09-06 11:23:18 | | | | |
| 3 | Camila Xu | + 100 word(s) | 1205 | 2021-09-06 11:24:11 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Cazzato, G. Artificial Intelligence in Dermatopathology. Encyclopedia. Available online: https://encyclopedia.pub/entry/13921 (accessed on 07 February 2026).
Cazzato G. Artificial Intelligence in Dermatopathology. Encyclopedia. Available at: https://encyclopedia.pub/entry/13921. Accessed February 07, 2026.
Cazzato, Gerardo. "Artificial Intelligence in Dermatopathology" Encyclopedia, https://encyclopedia.pub/entry/13921 (accessed February 07, 2026).
Cazzato, G. (2021, September 06). Artificial Intelligence in Dermatopathology. In Encyclopedia. https://encyclopedia.pub/entry/13921
Cazzato, Gerardo. "Artificial Intelligence in Dermatopathology." Encyclopedia. Web. 06 September, 2021.
Copy Citation
Artificial intelligence applied to pathological anatomyhas attracted a particular interest from pathologists and, in more detail, also from dermatopathologists. Increasing attention is being paid to the applications of AI and ML in the diagnosis of simple or more complex skin lesions, and the training of AI algorithms is gathering increasing feedback from the scientific community.
AI
machine learning
dermatopathology
skin
future
1. Introduction
In the last two decades, an unprecedented development of information technologies associated with a considerable increase in memory units has allowed giant strides to be made in the futuristic field of artificial intelligence (AI). Although this is the prerogative of the informatics and technological branches, the use of software and new technologies has spread to many different fields of medicine, indeed to practically all branches, including pathological anatomy and, therefore, also the subbranch of dermatopathology [1][2].
2. Specifics
Artificial intelligence applied to pathological anatomy [3] has attracted a particular interest from pathologists and, in more detail, also from dermatopathologists. Increasing attention is being paid to the applications of AI and ML in the diagnosis of simple or more complex skin lesions, and the training of AI algorithms is gathering increasing feedback from the scientific community [4][3][5][6]. In a recent paper by Sam Polesie et al. [7], an attempt was made to understand the degree of perception, attitude and knowledge of AI in general and then as applied to dermatology and dermatopathology. An anonymous, voluntary online survey was prepared and distributed to pathologists who regularly analyze dermatopathology slides/images. In total, 718 people from 91 countries responded to this survey (64.1% of them women). While 81.5% of the respondents were aware of AI as an emerging topic in pathology, only 18.8% had a good or excellent knowledge of AI. In terms of diagnosis classifications, 42.6% saw a strong or very strong potential for automatic suggestion of the possible skin cancer diagnosis. The corresponding figure for inflammatory skin diseases was 23.0%. For specific applications, the highest potential was considered to be for automatic detection of mitosis (79.2%) and tumor margins (62.1%) and for the evaluation of immunostaining (62.7%). The potential for automatic suggestion of immunostaining (37.6%) and genetic panels (48.3%) was seen as lower. This study highlighted that respondent age did not affect the general attitude towards AI. Only 6.0% of the respondents agreed or firmly agreed that human pathologists will be replaced by AI in the near future. Among the whole group, 72.3% agreed or firmly agreed that AI will improve dermatopathology and 84.1% think AI should be part of medical education. All this demonstrates profound recent changes in the very perception of AI and ML and that an increasing number of applications of these is occurring in various medical fields. For example, in some studies [8][9], artificial intelligence algorithms match or outperform doctors in disease detection related to medical imaging. Additionally, the use of AI has been facilitated by the availability of affordable high-speed Internet, new computing power and secure cloud storage to manage and share datasets. Therefore, it has been possible to make these algorithms scalable on multiple devices, platforms and operating systems, transforming them into modern medical tools [10]. The paper by Esteva et al. [11] applied a deep learning algorithm to a combined skin dataset of 129,450 clinical and dermoscopic images consisting of 2032 different skin lesions. The AI performance was shown to be on a par with the dermatologists’ performance for skin cancer classification. Deep learning solutions have been successful in the field of digital pathology with whole-slide imaging (WSI). Examples of histopathological images of skin lesions are shown in Figure 1.
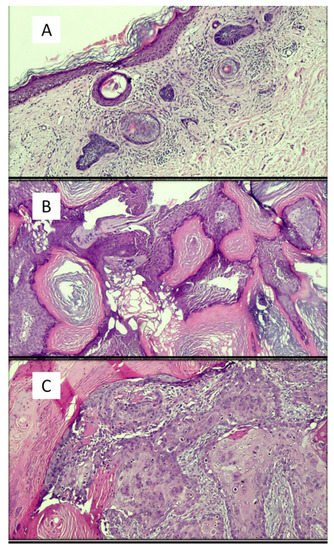
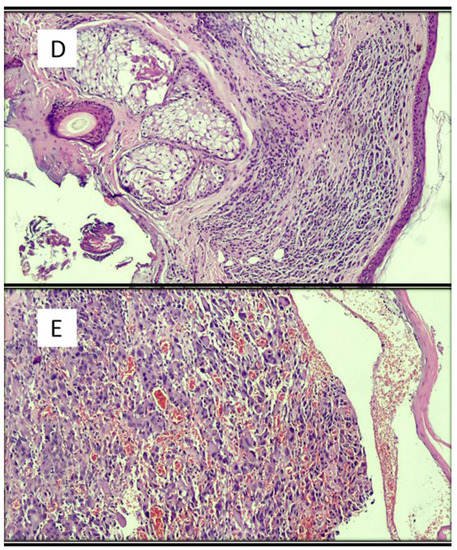
Figure 1. Examples of images of lesions used in various studies in the literature. (A) Basal cell carcinoma, superficial variant (hematoxylin–eosin, original magnification: 4×). (B) Seborrheic keratosis, hyperkeratotic variant (hematoxylin–eosin, original magnification: 10×). (C) Squamous cell carcinoma (hematoxylin–eosin, original magnification: 20×). (D) Intradermic nevus (hematoxylin–eosin, original magnification: 10×). (E) Amelanotic malignant melanoma (hematoxylin–eosin, original magnification: 20×).
Hekler et al. [12] analyzed 695 lesions previously classified by an expert histopathologist according to the guidelines (of which 350 were nevi and 345 were melanomas). Hematoxylin and eosin (H&E)-stained slides of these lesions were scanned using a slide scanner and then randomly cut out; 595 of the resulting images were used to train a convolutional neural network. The additional 100 sections of H&E images were used to test the CNN results against the original class labels. The authors reported a discrepancy with the histopathologist of 18% for melanoma (95% confidence interval (CI): 7.4–28.6%), 20% for nevi (95% CI: 8.9–31.1%) and 19% for the full set of images (95% CI: 11.3–26.7%).
Jiang et al. [13] aimed to develop deep neural network structures for accurate BCC recognition and segmentation based on microscopic ocular images (MOI) acquired by smartphones. To do this, they collected a total of 8046 MOIs, 6610 of which had binary classification labels, while the other 1436 had pixel-by-pixel annotations. Meanwhile, 128 WSIs were collected for comparison. Two deep learning frameworks were created. The “waterfall” framework had a classification model for identifying difficult cases (images with low prediction confidence) and a segmentation model for further in-depth analysis of difficult cases. The “segmentation” framework directly segmented and categorized all images.
Cruz-Roa et al. [14] used a deep learning architecture to discriminate between BCC and normal tissue models in 1417 images from 308 regions of interest (ROIs) of skin histopathology images. They compared the deep learning method to traditional machine learning with feature descriptors, including feature pack, canonical and Haar wavelet transformation. The deep learning architecture proved superior to traditional approaches, reaching 89.4% in F-measure and 91.4% in balanced accuracy.
Table 1 summarizes other studies present in the literature and not mentioned in the entry.
Table 1. Other studies present in the literature besides those analyzed in the Discussion section of this work.
| Authors | Years | Type of AI | Results | Strengths | Limits |
|---|---|---|---|---|---|
| Potter et al. [15] | 1987 | Interactive computer program |
Concordance, 91.8% Disagreement, 4.8% |
Concordance and possibility of integration with patient clinical data | Disagreement and little memory space |
| Crowlet R. et al. [16] | 2003 | Traditional intelligent tutoring system |
Possibility of learning rather easily | Positive feedback | Clear prototypical schemes are indispensable |
| Joset Feit et al. [17] | 2005 | Hypertext atlas of dermatopathology | A collection of about 3200 dermatopathological images | Continuous updating | / |
| Payne et al. [18] | 2009 | Intelligent tutoring system |
Tutoring made it possible to implement the training of learners | Ability to learn from mistakes | Greater difficulties in tutoring related to superficial perivascular dermatitis |
| Olsen et al. [19] | 2018 | Deep learning algorithms | The artificial intelligence system accurately classified 123/124 (99.45%) BCCs (nodular), 113/114 (99.4%) dermal nevi and 123/123 (100%) seborrheic keratoses | Concordance | Difficulty in presenting artifacts, poor coloring |
Peizhen et al. [20] built a multicenter database of 2241 digital images of whole slides of 1321 patients from 2008 to 2018. They trained both ResNet50 and Vgg19 using over 9.95 million patches by transferring learning and test performance using two types of critical classifications: melanoma malignant versus benign nevi in separate and mixed magnification, and distinguishing nevi at maximum magnification. Regions of interest (ROIs) were also localized, which was significantly useful, offering pathologists greater support for the correct diagnosis. Although the development of ML-based AI is spreading in dermatopathology, we are still quite far from its application in clinical routine because, even when compared to the algorithms applied in dermoscopy, there is less sensitivity and specificity and hence less accuracy. More specifically, some histological lesions closely mimic other types of neoplasms, such as skin adnexal lesions, which can require differential diagnosis with BCC, SCC, KS or melanoma [12]. Furthermore, despite rather promising values, these algorithms are not able to diagnose a malignant lesion (for example, melanoma) in all cases, thus making their use unacceptable without human control [12][13][14][19][20][21][22].
References
- Tizhoosh, H.R.; Pantanowitz, L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J. Pathol. Inform. 2018, 14, 38.
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88.
- Venerito, V.; Angelini, O.; Cazzato, G.; Lopalco, G.; Maiorano, E.; Cimmino, A.; Iannone, F. A convolutional neural network with transfer learning for automatic discrimination between low and high-grade synovitis: A pilot study. Intern Emerg. Med. 2021, 16, 1457–1465.
- Bauer, T.W.; Schoenfield, L.; Slaw, R.J.; Yerian, L.; Sun, Z.; Henricks, W.H. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch. Pathol. Lab. Med. 2013, 137, 518–524.
- Cazzato, G.; Colagrande, A.; Cimmino, A.; Liguori, G.; Lettini, T.; Serio, G.; Ingravallo, G.; Marzullo, A. Atypical Fibroxanthoma-Like Amelanotic Melanoma: A Diagnostic Challenge. Dermatopathology 2021, 12, 4.
- Onega, T.; Reisch, L.M.; Frederick, P.D.; Geller, B.M.; Nelson, H.D.; Lott, J.P.; Radick, A.C.; Elder, D.E.; Barnhill, R.L.; Piepkorn, M.W.; et al. Use of digital whole slide imaging in dermatopathology. J. Digit. Imaging 2016, 29, 243–253.
- Polesie, S.; McKee, P.H.; Gardner, J.M.; Gillstedt, M.; Siarov, J.; Neittaanmäki, N.; Paoli, J. Attitudes Toward Artificial Intelligence Within Dermatopathology: An International Online Survey. Front. Med. 2020, 20, 591952.
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297.
- Esteva, A.; Kuprel, B.; Novoa, N.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118.
- Bengio, Y.; Goodfellow, I.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016.
- Gomolin, A.; Netchiporouk, E.; Gniadecki, R.; Litvinov, I.V. Artificial Intelligence Applications in Dermatology: Where Do We Stand? Front. Med. (Lausanne) 2020, 31, 100.
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Berking, C.; Klode, J.; Schadendorf, D.; Jansen, P.; Franklin, C.; Holland-Letz, T.; Krahl, D.; et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur. J. Cancer 2019, 115, 79–83.
- Jiang, Y.Q.; Xiong, J.H.; Li, H.Y.; Yang, X.H.; Yu, W.T.; Gao, M.; Zhao, X.; Ma, Y.P.; Zhang, W.; Guan, Y.F.; et al. Recognizing basal cell carcinoma on smartphone-captured digital histopathology images with a deep neural network. Br. J. Dermatol. 2020, 182, 754–762.
- Cruz-Roa, A.A.; Arevalo Ovalle, J.E.; Madabhushi, A.; González Osorio, F.A. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med. Image Comput. Comput. Assist Interv. 2013, 16, 403–410.
- Potter, B.; Ronan, S.G. Computerized dermatopathologic diagnosis. J. Am. Acad. Dermatol. 1987, 17, 119–131.
- Crowley, R.S.; Medvedeva, O. A general architecture for intelligent tutoring of diagnostic classification problem solving. AMIA Annu. Symp. Proc. 2003, 2003, 185–189.
- Feit, J.; Kempf, W.; Jedlicková, H.; Burg, G. Hypertext atlas of dermatopathology with expert system for epithelial tumors of the skin. J. Cutan. Pathol. 2005, 32, 433–437.
- Payne, V.L.; Medvedeva, O.; Legowski, E.; Castine, M.; Tseytlin, E.; Jukic, D.; Crowley, R.S. Effect of a limited-enforcement intelligent tutoring system in dermatopathology on student errors, goals and solution paths. Artif. Intell. Med. 2009, 47, 175–197.
- Olsen, T.G.; Jackson, B.H.; Feeser, T.A.; Kent, M.N.; Moad, J.C.; Krishnamurthy, S.; Lunsford, D.D.; Soans, R.E. Diagnostic Performance of Deep Learning Algorithms Applied to Three Common Diagnoses in Dermatopathology. J. Pathol. Inform. 2018, 27, 32.
- Peizhen, I.; Xie, K.; Zuo, Y.; Zhang, F.; Li, M.; Yin, K. Interpretable Classification from Skin Cancer Histology Slides Using Deep Learning: A Retrospective Multicenter Study. arXiv 2019, arXiv:1904.06156.
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities. Comput. Biol. Med. 2020, 127, 104065.
- Cives, M.; Mannavola, F.; Lospalluti, L.; Sergi, M.C.; Cazzato, G.; Filoni, E.; Cavallo, F.; Giudice, G.; Stucci, L.S.; Porta, C.; et al. Non-Melanoma Skin Cancers: Biological and Clinical Features. Int. J. Mol. Sci. 2020, 21, 5394.
More
Information
Subjects:
Dermatology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
3 times
(View History)
Update Date:
06 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




