
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Patrick Matthias | + 2088 word(s) | 2088 | 2021-07-21 10:01:11 | | | |
| 2 | Lindsay Dong | Meta information modification | 2088 | 2021-09-02 05:58:35 | | |
Video Upload Options
Influenza is a zoonotic respiratory disease of major public health interest due to its pandemic potential, and a threat to animals and the human population. The influenza A virus genome consists of eight single-stranded RNA segments sequestered within a protein capsid and a lipid bilayer envelope. During host cell entry, cellular cues contribute to viral conformational changes that promote critical events such as fusion with late endosomes, capsid uncoating and viral genome release into the cytosol.
1. Introduction
Entry of an enveloped virus from the extracellular environment into cells proceeds through a number of essential steps [1]. These include binding and attachment of a virus outer protein to its receptor at the cell surface, penetration of the viral particle into the cytoplasm, uncoating of the proteinaceous capsid allowing release of the viral nucleic acids into the cell cytosol, viral genetic material replication, protein synthesis, and finally new viral particle assembly and budding from the infected cell. The dissection of the molecular events and viral–host interactions that take place once the virus binds to the cell surface is essential for understanding how a particular virus infects cells. It also allows the identification of potential new targets for antivirals and therapies for blocking or controlling the infection and onset of diseases.
As viruses recognize target cells by first binding to cell receptors, the discovery of the virus ligands is primordial for understanding the organ tropism, the potential host diversity, and the mechanism of infection [2][3]. Enveloped animal viruses enter their host cells by membrane fusion and two pathways have been described, depending on the characteristics of the virus fusion protein. Fusion can occur at the cell plasma membrane at physiological pH [4][5][6][7][8][9][10] or within the endocytic vacuolar system where it is triggered by a low pH [11][12][13][14][15][16][17]. Virus capsid opening, the so-called uncoating, enables the virus genetic material to be released in the cytosol and get ready for replication. Our understanding of virus uncoating mechanisms has grown substantially in recent years and defines uncoating as a complex, highly orchestrated, multi-step process that relies on both viral and cellular factors.
2. Influenza Virus Capsid Uncoating
2.1. Involvement of Ubiquitin Chains in Influenza Virus Uncoating
3.2. The Role of HDAC6 in Influenza Virus Uncoating
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl groups from modified lysine residues of histone and non-histone proteins and several classes of mammalian HDACs exist [26]. Class I HDACs are 400–500 amino acids long and include HDAC1, HDAC2, HDAC3 and HDAC8. Class II HDACs are approximately 1000 amino acids long; class IIa comprises HDAC4, HDAC5, HDAC7 and HDAC9, and class IIb comprises HDAC6 and HDAC10 [27][28][29]. The class III HDACs, also known as the sirtuins (SIRT1–7), are the silent information regulator 2 (Sir2) family of proteins and have a size ranging from 300 to 750 amino acids [30][31]. Despite the name, HDAC function is not limited to histone deacetylation and the regulation of gene transcription. HDAC6 localizes mainly in the cytosol and targets proteins through one of its deacetylase domains, CD1 or CD2, or its Ub-binding zinc finger domain, ZnF. In humans, HDAC6 contains a Ser Glu-repeat domain (SE14), which acts as a cytoplasmic retention signal and mediates its stable anchorage in the cytoplasm [32] where it deacetylases tubulin [33][34][35], heat shock protein 90 (Hsp90) [36][37][38], β-catenin [39][40], cortactin [41], MYH9, Hsc70, DNAJA1 [42] or the DEAD box RNA helicase 3, X-linked (DDX3X) [43]. HDAC6 has been associated with carcinogenesis, neurodegenerative diseases and inflammatory disorders, and has been exploited as a therapeutic target for pharmacological intervention [44][45][46][47][48][49][50][51][52].
HDAC6 has also been identified to have antiviral effects which have been linked to its enzymatic activity. It was found to inhibit IAV release by downregulating the trafficking of viral components to the plasma membrane via acetylated microtubules [53]. Overexpression of HDAC6 in cells leads to diminished viral budding due to tubulin deacetylation [54].
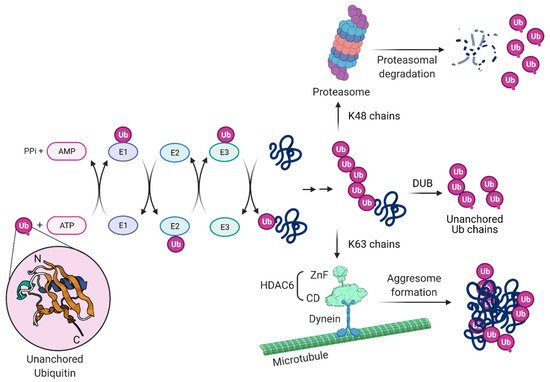
HDAC6, through its ZnF domain, associates with unanchored Ub chains [55][56][57]. As shown in Figure 3, Ub chains can be generated through E1/E2/E3 cycles and unanchored Ub chains are present in monoubiquitin, or ubiquitin derived from proteasomal degradation or the catalytic action of DUBs on pre-existing Ub chains. HDAC6 ZnF binds Ub with high affinity by recognizing its unanchored C-terminal sequence (-RLRGG-COOH) [58][59] and can recruit misfolded proteins with entangled Ub chains.
Inflammasome complexes are formed in response to pathogen-associated molecules and, for NLR family pyrin domain-containing protein 3 (NLRP3)- and pyrin-mediated inflammasomes, their assembly and downstream functions occur at the MTOC [60]. Similar to the formation of aggresomes, HDAC6 ZnF is required for the interaction with NLRP3 and pyrin inflammasome components and transport of these proteins using the microtubule retrograde transport by dynein for their activation in macrophages [60].
2.3. Epidermal Growth Factor Receptor Pathway Substrate 8
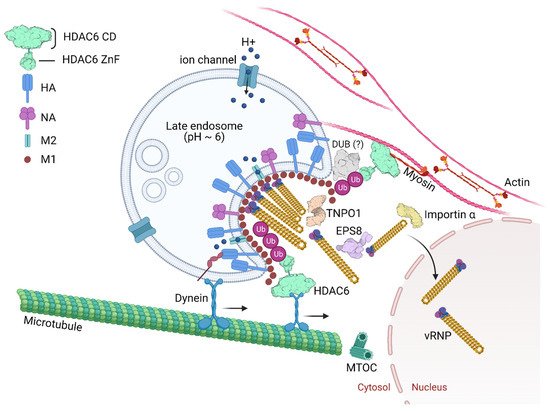
The epidermal growth factor receptor pathway substrate 8 (EPS8) is an adaptor protein involved in signaling via the epidermal growth factor receptor (EGFR) [61][62]. EPS8 physically associates with incoming virus components possibly through interactions with NP, the viral polymerase, M1, or bridged by other cellular uncoating factors (Figure 4). EPS8 might interact with vRNPs by binding to NP as the viral nucleoprotein specifically co-precipitated with EPS8.
2.4. SPOPL/Cullin 3 Ubiquitin Ligase Complex and EPS15
The maturation of late endosomes/multivesicular bodies entails the spatial and functional separation of the organelles from early endosomes, preparing them as a feeder pathway to lysosomes [63]. Cellular processes that promote endosome maturation play a critical role in influenza uncoating. Cullin3 (CUL3)-based E3 ubiquitin ligases regulate endocytic trafficking of cargo to lysosomes and endosome maturation. Transfer of cargo from early endosomes to lysosomes depends on an endosomal maturation process regulated by a variety of protein- and lipid-based events.
2.5. Transportin 1
Importins mediate the nuclear import of cargos and transportin 1 (TNPO1, also known as importin-β2, KPNB2) is one of the best-characterized nuclear import receptors [64].
Many viruses depend on nuclear proteins for replication and their viral genome must enter the nucleus of the host cell. This is the case of most DNA viruses and some RNA viruses, including orthomyxoviruses and retroviruses. Therefore, it is expected that the life cycle of these viruses is dependent on transporters (e.g., importins, exportins, transportins) and regulators (e.g., Ran GTPase). Great effort has been put on deciphering viral nuclear transport mechanisms [65][66][67][68]. In the context of IAV infection, a study using RNAi for targeting, among other proteins, nuclear pore proteins identified TNPO1 as an important host factor involved in uncoating. Depletion of TNPO1 in different cells reduced the number of infected cells and the production of new viruses [69]. It was shown that TNPO1 was important not only for the vRNPs nuclear import, but also for the M1 uncoating and vRNP debundling in the cytosol [69]. Moreover, the role of TNPO1 in uncoating was associated with its recognition of a nuclear localization signal as it binds to the exposed M1 N-terminal PY-NLS motif only after capsid acidification. As shown in Figure 2, by recognizing and binding the M1 NLS, TNPO1 promotes the removal of vRNP-associated M1, which leads to dissociation of vRNPs from each other and facilitates further nuclear import by importin α and β via the classical NLS-mediated import pathway.
References
- Ryu, W.-S. Virus Life Cycle. Mol. Virol. Hum. Pathog. Viruses 2017.
- Ashok, A.; Atwood, W.J. Virus Receptors and Tropism. In Polyomaviruses and Human Diseases; Ahsan, N., Ed.; Springer: New York, NY, USA, 2006; pp. 60–72.
- Baranowski, E.; Ruiz-Jarabo, C.M.; Domingo, E. Evolution of Cell Recognition by Viruses. Science 2001, 292, 1102.
- Greenstone, H.L.; Santoro, F.; Lusso, P.; Berger, E.A. Human Herpesvirus 6 and Measles Virus Employ Distinct CD46 Domains for Receptor Function. J. Biol. Chem. 2002, 277, 39112–39118.
- Spear, P.G.; Eisenberg, R.J.; Cohen, G.H. Three Classes of Cell Surface Receptors for Alphaherpesvirus Entry. Virology 2000, 275, 1–8.
- Morgan, C.; Rose, H.M.; Mednis, B. Electron Microscopy of Herpes Simplex Virus: I. Entry. J. Virol. 1968, 2, 507–516.
- Fuller, A.O.; Spear, P.G. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. USA 1987, 84, 5454–5458.
- Wittels, M.; Spear, P.G. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 1991, 18, 271–290.
- Aguilar, H.C.; Henderson, B.A.; Zamora, J.L.; Johnston, G.P. Paramyxovirus Glycoproteins and the Membrane Fusion Process. Curr. Clin. Microbiol. Rep. 2016, 3, 142–154.
- Chauveau, L.; Donahue, D.A.; Monel, B.; Porrot, F.; Bruel, T.; Richard, L.; Casartelli, N.; Schwartz, O. HIV Fusion in Dendritic Cells Occurs Mainly at the Surface and Is Limited by Low CD4 Levels. J. Virol. 2017, 91, e01248-17.
- Akula, S.M.; Naranatt, P.P.; Walia, N.-S.; Wang, F.-Z.; Fegley, B.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus (Human Herpesvirus 8) Infection of Human Fibroblast Cells Occurs through Endocytosis. J. Virol. 2003, 77, 7978–7990.
- Acosta, E.G.; Castilla, V.; Damonte, E.B. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J. Gen. Virol. 2008, 89, 474–484.
- Persaud, M.; Martinez-Lopez, A.; Buffone, C.; Porcelli, S.A.; Diaz-Griffero, F. Infection by Zika viruses requires the transmembrane protein AXL, endocytosis and low pH. Virology 2018, 518, 301–312.
- Doms, R.W.; Helenius, A.; White, J. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 1985, 260, 2973–2981.
- Ruigrok, R.W.H.; Aitken, A.; Calder, L.J.; Martin, S.R.; Skehel, J.J.; Wharton, S.A.; Weis, W.; Wiley, D.C. Studies on the Structure of the Influenza Virus Haemagglutinin at the pH of Membrane Fusion. J. Gen. Virol. 1988, 69, 2785–2795.
- Aleksandrowicz, P.; Marzi, A.; Biedenkopf, N.; Beimforde, N.; Becker, S.; Hoenen, T.; Feldmann, H.; Schnittler, H.-J. Ebola Virus Enters Host Cells by Macropinocytosis and Clathrin-Mediated Endocytosis. J. Infect. Dis. 2011, 204, S957–S967.
- Li, L.; Jose, J.; Xiang, Y.; Kuhn, R.J.; Rossmann, M.G. Structural changes of envelope proteins during alphavirus fusion. Nature 2010, 468, 705–708.
- Banerjee, I.; Miyake, Y.; Nobs, S.P.; Schneider, C.; Horvath, P.; Kopf, M.; Matthias, P.; Helenius, A.; Yamauchi, Y. Influenza A virus uses the aggresome processing machinery for host cell entry. Science 2014, 346, 473–477.
- Rudnicka, A.; Yamauchi, Y. Ubiquitin in Influenza Virus Entry and Innate Immunity. Viruses 2016, 8, 293.
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533.
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440.
- Dikic, I.; Dötsch, V. Ubiquitin linkages make a difference. Nat. Struct. Mol. Biol. 2009, 16, 1209–1210.
- Miyake, Y.; Matthias, P.; Yamauchi, Y. Purification of Unanchored Polyubiquitin Chains from Influenza Virions. In Influenza Virus: Methods and Protocols; Yamauchi, Y., Ed.; Springer: New York, NY, USA, 2018; pp. 329–342.
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.S.; Ito, A.; Yoshida, M.; Yao, T.-P. Proteasomes Activate Aggresome Disassembly and Clearance by Producing Unanchored Ubiquitin Chains. Mol. Cell 2013, 51, 819–828.
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407.
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713.
- Schultz, B.E.; Misialek, S.; Wu, J.; Tang, J.; Conn, M.T.; Tahilramani, R.; Wong, L. Kinetics and Comparative Reactivity of Human Class I and Class IIb Histone Deacetylases. Biochemistry 2004, 43, 11083–11091.
- Verdin, E.; Dequiedt, F.; Kasler, H.G. Class II histone deacetylases: Versatile regulators. Trends Genet. 2003, 19, 286–293.
- Yang, X.-J.; Grégoire, S. Class II Histone Deacetylases: From Sequence to Function, Regulation, and Clinical Implication. Mol. Cell Biol. 2005, 25, 2873–2884.
- Marks, P.A.; Miller, T.; Richon, V.M. Histone deacetylases. Curr. Opin. Pharmacol. 2003, 3, 344–351.
- Liu, T.; Liu, P.Y.; Marshall, G.M. The Critical Role of the Class III Histone Deacetylase SIRT1 in Cancer. Cancer Res. 2009, 69, 1702–1705.
- Bertos, N.R.; Gilquin, B.; Chan, G.K.; Yen, T.J.; Khochbin, S.; Yang, X.J. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J. Biol. Chem. 2004, 279, 48246–48254.
- Zhang, Y.; Li, N.; Caron, C.; Matthias, G.; Hess, D.; Khochbin, S.; Matthias, P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003, 22, 1168–1179.
- Matsuyama, A.; Shimazu, T.; Sumida, Y.; Saito, A.; Yoshimatsu, Y.; Seigneurin-Berny, D.; Osada, H.; Komatsu, Y.; Nishino, N.; Khochbin, S.; et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 2002, 21, 6820–6831.
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458.
- Bali, P.; Pranpat, M.; Bradner, J.; Balasis, M.; Fiskus, W.; Guo, F.; Rocha, K.; Kumaraswamy, S.; Boyapalle, S.; Atadja, P.; et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 2005, 280, 26729–26734.
- Kovacs, J.J.; Murphy, P.J.; Gaillard, S.; Zhao, X.; Wu, J.T.; Nicchitta, C.V.; Yoshida, M.; Toft, D.O.; Pratt, W.B.; Yao, T.P. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 2005, 18, 601–607.
- Murphy, P.J.; Morishima, Y.; Kovacs, J.J.; Yao, T.P.; Pratt, W.B. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J. Biol. Chem. 2005, 280, 33792–33799.
- Li, Y.; Zhang, X.; Polakiewicz, R.D.; Yao, T.P.; Comb, M.J. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J. Biol. Chem. 2008, 283, 12686–12690.
- Wang, S.; Li, N.; Wei, Y.; Li, Q.; Yu, Z. β-catenin deacetylation is essential for WNT-induced proliferation of breast cancer cells. Mol. Med. Rep. 2014, 9, 973–978.
- Zhang, X.; Yuan, Z.; Zhang, Y.; Yong, S.; Salas-Burgos, A.; Koomen, J.; Olashaw, N.; Parsons, J.T.; Yang, X.J.; Dent, S.R.; et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 2007, 27, 197–213.
- Zhang, L.; Liu, S.; Liu, N.; Zhang, Y.; Liu, M.; Li, D.; Seto, E.; Yao, T.P.; Shui, W.; Zhou, J. Proteomic identification and functional characterization of MYH9, Hsc70, and DNAJA1 as novel substrates of HDAC6 deacetylase activity. Protein Cell 2015, 6, 42–54.
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019, 15, 51–61.
- Dallavalle, S.; Pisano, C.; Zunino, F. Development and therapeutic impact of HDAC6-selective inhibitors. Biochem. Pharmacol. 2012, 84, 756–765.
- Krämer, O.H.; Mahboobi, S.; Sellmer, A. Drugging the HDAC6–HSP90 interplay in malignant cells. Trends Pharmacol. Sci. 2014, 35, 501–509.
- Butler, K.V.; Kalin, J.; Brochier, C.; Vistoli, G.; Langley, B.; Kozikowski, A.P. Rational Design and Simple Chemistry Yield a Superior, Neuroprotective HDAC6 Inhibitor, Tubastatin A. J. Am. Chem. Soc. 2010, 132, 10842–10846.
- Ma, J.; Huo, X.; Jarpe, M.B.; Kavelaars, A.; Heijnen, C.J. Pharmacological inhibition of HDAC6 reverses cognitive impairment and tau pathology as a result of cisplatin treatment. Acta Neuropathol. Commun. 2018, 6, 103.
- Gupta, R.; Ambasta, R.K.; Kumar, P. Pharmacological intervention of histone deacetylase enzymes in the neurodegenerative disorders. Life Sci. 2020, 243, 117278.
- LoPresti, P. HDAC6 in Diseases of Cognition and of Neurons. Cells 2021, 10, 12.
- Ganai, S.A. Small-molecule Modulation of HDAC6 Activity: The Propitious Therapeutic Strategy to Vanquish Neurodegenerative Disorders. Curr. Med. Chem. 2017, 24, 4104–4120.
- Pandey, D.; Nomura, Y.; Rossberg, M.; Bhatta, A.; Romer, L.; Berkowitz, D. A5942 Inhibition of Histone Deacetylase 6 activity provides protection against atherogenesis: A role for HDAC6 NEDDylation. J. Hypertens. 2018, 36, e44–e45.
- Prior, R.; Van Helleputte, L.; Klingl, Y.E.; Van Den Bosch, L. HDAC6 as a potential therapeutic target for peripheral nerve disorders. Expert Opin. Ther. Targets 2018, 22, 993–1007.
- Husain, M.; Cheung, C.Y. Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J. Virol. 2014, 88, 11229–11239.
- Husain, M.; Harrod, K.S. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett. 2011, 585, 128–132.
- Hook, S.S.; Orian, A.; Cowley, S.M.; Eisenman, R.N. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 2002, 99, 13425–13430.
- Seigneurin-Berny, D.; Verdel, A.; Curtet, S.; Lemercier, C.; Garin, J.; Rousseaux, S.; Khochbin, S. Identification of components of the murine histone deacetylase 6 complex: Link between acetylation and ubiquitination signaling pathways. Mol. Cell Biol. 2001, 21, 8035–8044.
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003, 115, 727–738.
- Dos Santos Passos, C.; Simões-Pires, C.A.; Carrupt, P.A.; Nurisso, A. Molecular dynamics of zinc-finger ubiquitin binding domains: A comparative study of histone deacetylase 6 and ubiquitin-specific protease 5. J. Biomol. Struct. Dyn. 2016, 34, 2581–2598.
- Hard, R.L.; Liu, J.; Shen, J.; Zhou, P.; Pei, D. HDAC6 and Ubp-M BUZ domains recognize specific C-terminal sequences of proteins. Biochemistry 2010, 49, 10737–10746.
- Magupalli, V.G.; Negro, R.; Tian, Y.; Hauenstein, A.V.; Di Caprio, G.; Skillern, W.; Deng, Q.; Orning, P.; Alam, H.B.; Maliga, Z.; et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science 2020, 369, eaas8995.
- Fazioli, F.; Minichiello, L.; Matoska, V.; Castagnino, P.; Miki, T.; Wong, W.T.; Di Fiore, P.P. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993, 12, 3799–3808.
- Lanzetti, L.; Rybin, V.; Malabarba, M.G.; Christoforidis, S.; Scita, G.; Zerial, M.; Di Fiore, P.P. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 2000, 408, 374–377.
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500.
- Mboukou, A.; Rajendra, V.; Kleinova, R.; Tisné, C.; Jantsch, M.F.; Barraud, P. Transportin-1: A Nuclear Import Receptor with Moonlighting Functions. Front. Mol. Biosci. 2021, 8, 638149.
- Flatt, J.W.; Greber, U.F. Viral mechanisms for docking and delivering at nuclear pore complexes. Semin. Cell Dev. Biol. 2017, 68, 59–71.
- Greber, U.F.; Fornerod, M. Nuclear Import in Viral Infections. In Membrane Trafficking in Viral Replication; Marsh, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 109–138.
- Yarbrough, M.L.; Mata, M.A.; Sakthivel, R.; Fontoura, B.M.A. Viral Subversion of Nucleocytoplasmic Trafficking. Traffic 2014, 15, 127–140.
- Fontoura, B.M.A.; Faria, P.A.; Nussenzveig, D.R. Viral Interactions with the Nuclear Transport Machinery: Discovering and Disrupting Pathways. IUBMB Life 2005, 57, 65–72.
- Miyake, Y.; Keusch, J.J.; Decamps, L.; Ho-Xuan, H.; Iketani, S.; Gut, H.; Kutay, U.; Helenius, A.; Yamauchi, Y. Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat. Microbiol. 2019, 4, 578–586.




