
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | beata smolarz | + 2009 word(s) | 2009 | 2021-08-10 11:36:02 | | | |
| 2 | Lindsay Dong | Meta information modification | 2009 | 2021-09-02 06:16:20 | | |
Video Upload Options
Long noncoding RNAs (lncRNAs) are the largest groups of ribonucleic acids, but, despite the increasing amount of literature data, the least understood. Given the involvement of lncRNA in basic cellular processes, especially in the regulation of transcription, the role of these noncoding molecules seems to be of great importance for the proper functioning of the organism. Studies have shown a relationship between disturbed lncRNA expression and the pathogenesis of many diseases, including cancer.
1. lncRNA—History
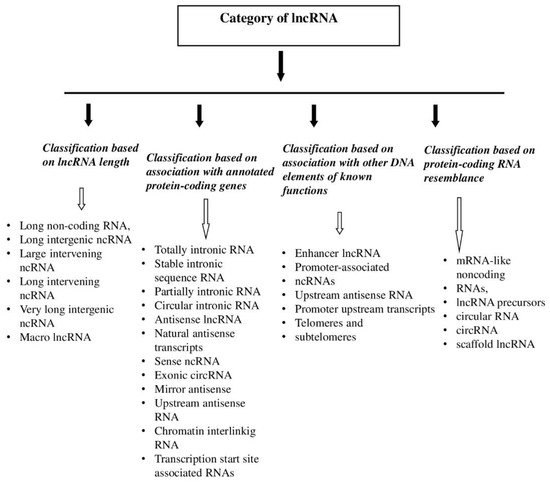
2. lncRNA—Characteristics
3. lncRNA—Functions
lncRNA affects the transcription process directly, acting as an enhancer, by “stopping” transcription factors, and by affecting chromatin looping and gene methylation (using epigenetic complexes such as PCR2) [9].
4. lncRNA and Malignant Tumors
The characteristics of long noncoded RNA described above—such as tissue or cellular specificity and the regulation of gene expression at the transcriptional and post-transcriptional levels—indicate that lncRNAs may be important in the formation of malignant tumors. Studies have shown that lncRNAs affect the pathways of division, growth, and cell differentiation, and are also involved in cellular death processes [9][11]. Modifications to these processes may lead to carcinogenesis [11]. Moreover, some lncRNAs are regulated by oncogene products or cancer transformation suppressors, which means that they are believed to indirectly perform tumorigenic functions (Table 1) [11].
| lncRNA | Genomic Location | Expression in Patients | Function in Tumorigenesis |
|---|---|---|---|
| PCGEM1 | 2q32.2 | Increased in prostate cancer | oncogene |
| MALAT1 | 11q13.1 | Increased in colon, lung, and liver cancers | oncogene |
| MEG3 | 14q32.2 | Down-regulated in multiple cancers | tumor suppressor |
| HOTAIR | 12q13.13 | Increased in primary breast tumors and metastases, GIST, and pancreatic cancers |
oncogene |
The first observed transcripts of altered expression in the tissue of a malignant tumor—prostate cancer—were PCA3 and PCGEM1. PCA3 currently functions as a cancer marker [11]. The previously mentioned MALAT1 was also discovered in cancerous tissue as one of the first lncRNAs. Of prognostic significance, its altered expression was discovered in the tissues of lung cancer [12]. It is now known that altered expression of this transcript occurs in many cancers, which may indicate its importance in the process of cellular proliferation [13].
5. lncRNA—T-UCRs
6. T-UCRs and Malignant Tumors
7. Conclusive Summary
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Inter-national human genome sequencing consortium. Initial sequencing and analysis of the human ge-nome. Nature 2001, 409, 860–921.
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351.
- Jarroux, J.; Morillon, A.; Pinskaya, M. Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46.
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36.
- XIST Gene—GeneCards|XIST RNA Gene. Weizmann Institute of Science. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=XIST&keywords=xist (accessed on 4 August 2021).
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373.
- Ayupe, A.C.; Tahira, A.C.; Camargo, L.; Beckedorff, F.C.; Verjovski-Almeida, S.; Reis, E.M. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic re-gions of the human genome. RNA Biol. 2015, 12, 877–892.
- Giannakakis, A.; Zhang, J.; Jenjaroenpun, P.; Nama, S.; Zainolabidin, N.; Aau, M.Y.; Yarmishyn, A.A.; Vaz, C.; Ivshina, A.V.; Grinchuk, O.V.; et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci. Rep. 2015, 5, 9737.
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463.
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407.
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261.
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041.
- Gutschner, T.; Hämmerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801.
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366.
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629.
- Léveillé, N.; Melo, C.A.; Rooijers, K.; Díaz-Lagares, A.; Melo, S.A.; Korkmaz, G.; Lopes, R.; Moqadam, F.A.; Maia, A.R.; Wijchers, P.J.; et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015, 6, 6520.
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742.
- Kim, T.; Jeon, Y.J.; Cui, R.; Lee, J.H.; Peng, Y.; Kim, S.H.; Tili, E.; Alder, H.; Croce, C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015, 107, 4.
- Hart, J.R.; Roberts, T.C.; Weinberg, M.S.; Morris, K.V.; Vogt, P.K. MYC regulates the non-coding transcriptome. Oncotarget 2014, 5, 12543–12554.
- Sánchez, Y.; Segura, V.; Marín-Béjar, O.; Athie, A.; Marchese, F.P.; González, J.; Bujanda, L.; Guo, S.; Matheu, A.; Huarte, M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014, 5, 5812.
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27.
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419.
- Jiang, R.; Tang, J.; Chen, Y.; Deng, L.; Ji, J.; Xie, Y.; Wang, K.; Jia, W.; Chu, W.M.; Sun, B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellu-lar carcinoma immune evasion. Nat. Commun. 2017, 8, 15129.
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultracon-served elements in the human genome. Science 2004, 304, 1321–1325.
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229.
- Pasmant, E.; Sabbagh, A.; Vidaud, M.; Bièche, I. ANRIL, a long, noncoding RNA, is an unex-pected major hotspot in GWAS. FASEB J. 2011, 25, 444–448.
- Terracciano, D.; Terreri, S.; de Nigris, F.; Costa, V.; Calin, G.A.; Cimmino, A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochimica et biophysica acta. Rev. Cancer 2017, 1868, 449–455.
- Ferdin, J.; Nishida, N.; Wu, X.; Nicoloso, M.S.; Shah, M.Y.; Devlin, C.; Ling, H.; Shimizu, M.; Kumar, K.; Cortez, M.A.; et al. HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved tran-scripts. Cell Death Differ. 2013, 20, 1675–1687.
- Mestdagh, P.; Fredlund, E.; Pattyn, F.; Rihani, A.; Van Maerken, T.; Vermeulen, J.; Kumps, C.; Menten, B.; De Preter, K.; Schramm, A.; et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010, 29, 3583–3592.
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606.
- Andrews, S.J.; Rothnagel, J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014, 15, 193–204.
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22.




