| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sidrit Uruci | + 3378 word(s) | 3378 | 2021-08-27 08:44:28 | | | |
| 2 | Dean Liu | Meta information modification | 3378 | 2021-09-02 05:16:02 | | |
Video Upload Options
R-loops have been associated with both physiological and pathological functions that are conserved across species. R-loops are a source of replication stress and genome instability, as seen in neurodegenerative disorders and cancer. In response, cells have evolved pathways to prevent R-loop accumulation as well as to resolve them. This review covers various mechanisms precluding R‐loop accumulation and highlights the role of Chro-Mates (chromatin modifiers and remodelers) in facilitating timely R‐loop resolution. We also discuss the enigmatic role of RNA:DNA hybrids in facilitating DNA repair, epigenetic landscape and the potential role of replication fork preservation pathways, active fork stability and stalled fork protection pathways, in avoiding replication-transcription conflicts.
1. Introduction
The unwinding of the DNA double helix during events such as transcription, DNA replication or DNA repair, offers the opportunity for various anomalies, such as RNA:DNA hybrids or R-loops to form. R-loops were first discovered more than 40 years ago [1] and were named as “R-loops” to depict the three-stranded structure similar to previously described D-loops [1], but with an RNA moiety in the hybrid. It is still not fully understood how R-loops are generated. The most popular model, the “thread back model”, is supported by crystallographic analysis [2][3] and suggests that the newly formed RNA strand threads back between the two DNA strands before they reanneal, forming the RNA:DNA hybrid and displacing the non-template single-strand DNA [4][5]. Initially, R-loops were thought to be transient and to be produced only as a byproduct of transcription, but their greater significance is now coming to light [6][7][8][9]. The process of R-loop formation is thermodynamically favored since the RNA:DNA hybrid interaction is much stronger and more stable than that of DNA:DNA duplexes [10][11]. There are many theories as to why this may be the case and, in addition to GC skew (the asymmetric distribution of G and C residues along the two strands), negative supercoils and DNA nicks are believed to play a fundamental role in RNA:DNA interaction and thus in enhancing R-loop stability. Deep-sequencing techniques have revealed that about 60% of the human transcribed genome contains at least one R-loop Forming Sequence (RLFS) [12]. Furthermore, R-loops have been observed in the genomes of organisms ranging from bacteria [13][14] to yeast [15][16], plants [17][18] and mammals [19][20]. In human, R-loops are formed at a modest frequency that ranges from 0.5% to 10% along tens of thousands of highly conserved hotspots [20][21]. R-loops’ roles are often divided into two main categories: “scheduled” R-loops, which are involved in normal physiological pathways of the cell cycle, and “unscheduled” R-loops, that appear to form only during episodes of cellular dysregulation and have been linked to replication stress, DNA damage, and to several human pathologies such as neurodegenerative diseases and cancer [7][9]
2. Prevention Mechanisms to Avoid R-loops Accumulation
Persistent and unhindered R-loops pose a threat to genome stability, therefore, it is fundamental for cells to maintain a homeostasis of R-loop abundance and to prevent their uncontrolled accumulation [9]. This regulation is crucial to maintain the roles of R-loops in their physiological processes and, at the same time, to minimize their pathological impact on genome stability. Alteration of this homeostasis is a major driver of genome and chromosome instability, hallmarks of oncogenesis and neurodegenerative diseases [51,63]. The prevention mechanisms largely act at the level of transcript regulation. Early investigations in budding yeast showed that the perturbation of mRNA biogenesis at any level promotes R-loop formation and DNA damage, thus the natural processing of transcripts acts as a first line of defense against unscheduled R-loop formation [79].
3. Resolving Mechanisms to Remove R-Loops
When regulatory mechanisms fail to prevent R-loop accumulation, the excess R-loops require resolution pathways for their removal. The nuclease activity of Ribonuclease H (RNase H1 and RNase H2), enzymes that are highly conserved from bacteria through metazoans and mammals, in degrading the RNA in RNA:DNA hybrids is crucial to this process [22]. Both enzymes share the essential Hybrid Binding Domain (HBD) through which they bind the RNA:DNA hybrid and degrade the RNA moiety via endonuclease activity. RNase H1 acts on R-loops formed during transcription and plays a role in transcription termination by degrading RNA:DNA hybrids at G-rich pause sites located downstream of the poly(A) site and behind the elongating RNAPII [37]. RNase H2 however, can recognize and cleave misincorporated ribonucleotides in the DNA duplex as wells as remove RNA primers within Okazaki fragments during DNA synthesis [25,99]. While RNase H2 can process R-loops created during DNA replication and repair and is possibly cell cycle regulated, as suggested in yeast, RNase H1 can function independently of the cell cycle to remove R-loops and appears to become activated in response to high R-loops loads [23].
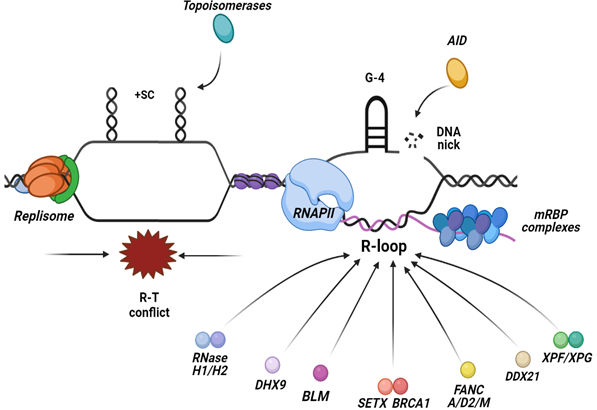
The R-loops can also be actively processed into DSBs by factors of the Transcription-Coupled Nucleotide Excision Repair (TC-NER) pathway. The fact that R-loops are processed by many distinct mechanisms, it is likely that R-loops form at any stage of the cell cycle. However, to preclude their accumulation, cells use other conserved mechanisms that are normally active during a specific cell cycle stage. Possibly, TC-NER factors have a role in recognizing and resolving R-loops associated with paused transcription, mainly in G1 cells. Similarly, there are other factors and pathways that exist during DNA replication in the S phase of the cell cycle, ensuring the resolution of frequent R-T conflicts, mostly through the unwinding activity of RNA/DNA helicases. Among several helicases, there are a few that have relevance in resolving R-loops and discussed below, such as the DEAD-box family helicases DDX21 [110], DHX9 helicase [111], BLM helicase [112], the helicases from the Fanconi Anemia (FA) pathway [113] and the human SETX helicase [56,114] (Figure 2).
Figure 2. R-loops are formed at the site of collisions between the replisome and RNAPII when they move along the DNA in opposite directions, leading to a head-on conflict between the two machineries. The main preventative mechanisms that inhibit R-loops accumulation are the regulation of torsional stress via the topoisomerases activity that relaxes supercoiled DNA in the vicinity of the forks and the transcript regulation, ensuring the proper packaging and processing of the mRNA. Factors implicated in the resolution of R-loops include nucleases such as, RNase H1/2 enzymes that hydrolyze the RNA moiety of the RNA:DNA hybrid and TC-NER factors, and a plethora of RNA/DNA helicases that unwind the hybrid.
The Enigmatic Role of RNA:DNA Hybrids in DNA Repair
For decades, unscheduled formation of R-loop/RNA:DNA hybrids has been considered as cause of DNA damage and genome instability. However, recent studies have suggested a rather fascinating role for transient RNA:DNA hybrids formation at sites of DNA damage in facilitating DNA repair. A recent study reports an interaction between BRCA2 and DDX5, a known DEAD-box helicase that also regulates resolution of RNA:DNA hybrids [139], particularly at DSBs sites. It is shown that BRCA2 stimulates the RNA:DNA helicase activity of DDX5 favoring its association with RNA:DNA hybrids in the vicinity of DSBs and finally promoting Homologous Recombination (HR) repair pathway [140]. Such role for BRCA2 has also been shown in another study that shows how BRCA2 regulates RNA:DNA hybrids levels at the DSBs site by mediating RNase H2 recruitment during S/G2 cell-cycle phase [141]. Moreover, it has been suggested that microRNA (miRNA) biogenesis enzymes, DROSHA and DICER, control the recruitment of repair factors from multiple pathways to sites of damage. DROSHA is suggested to be required for RNA processing within minutes of break induction, thus playing a central role in early stages of DNA repair. In the absence of DROSHA, a significant reduction of DNA repair by both HR and NHEJ was reported [142].
The role of RNA:DNA hybrids in DNA repair was also observed in fission yeast, where deletion of RNase H1 and RNase H2 resulted in the accumulation of RNAPII and RNA:DNA hybrids at the site of DSBs, leading to the inhibition of HR-mediated DSB repair [143]. Similarly, a recent study in budding yeast has associated the Sen1 ortholog with DSBs repair, as previously proposed in human [144]. Cells lacking a functional Sen1 present elevated levels of persistent RNA:DNA hybrids in the proximity of DSBs. At these DSB sites, RNA:DNA hybrids in concert with DNA nucleases Mre11 and Dna2, initiate DSB-resection through a non-canonical mechanism. However, persistence of RNA:DNA hybrids at the DSB site might also interfere with the resection process, leading to increased activation of mutagenic repair pathways such as NHEJ and Micro-homology-Mediated End Joining (MMEJ) [35]. Moreover, the role of R-loops has also been associated with the checkpoint activation. These mechanisms are detailed in a recent review article [62]. Briefly, the CD conflicts were associated with Ataxia Telangectasia Mutated (ATM) pathway activation while HO conflicts were associated with Ataxia Telangiectasia and Rad3-related protein (ATR) pathway activation, although the nature of this distinction is still unknown [58]. However, a recent study has further shown that loss of ATR, but not ATM, leads to significant increase in R-loop levels that are the ultimate source of replication stress and thus the ATR pathway being primarily required for their resolution [148]. However, this study also implicates that the slight increase in R-loop levels observed upon loss of ATM is possibly a consequence of unrepaired DSBs that accumulate genome wide in absence of ATM rather than a cause of DSBs formation.
4. Role of Epigenetic Marks in R-loop Homeostasis
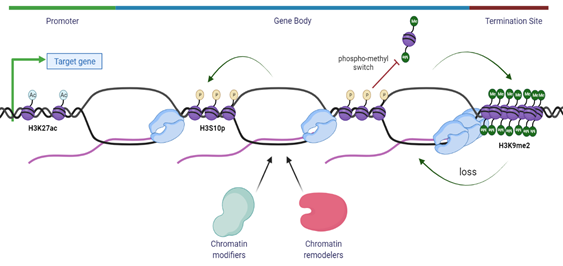
An intriguing idea that R-loops may act as an epigenetic layer has also emerged from the fact that R-loops show co-occurrence with specific chromatin epigenetic marks and post-translational modifications (PTMs), in mapped R-loops loci [149,150]. As previously discussed, R-loops are frequently found at unmethylated CpG island promoters [11] and at the 3′-end of several genes [22] where they play a key role in transcription termination [37]. Using an innovative R-loop genome-wide mapping method, NA:RNA Immuno-Precipitation followed by cDNA conversion coupled to high-throughput sequencing (DRIPc-Seq), R-loops have been shown to associate with specific epigenomic marks at promoters and terminators [23]. At the promoters, R-loops are enriched in histone marks such as methylation of H3, as seen with mono- and tri- methylation of lysine 4 and 36 residues of histone 3 (H3K4me1/3, H3K36me3) and open chromatin marks of histone 3 acetylation, such as the acetylation of lysine 27 residue (H3K27ac), while at terminators, R-loops are associated with H3K4me1, suggesting that H3K4me1 is a common mark of R-loop regions [11,22,23,39]. Even though all these epigenetic marks are associated with increased chromosome accessibility and thus active gene expression, it is intriguing how chromatin modifiers may sense the chromatin status associated with distinct R-loops that further play a role in epigenetic regulation on gene expression [39] (Figure 3).
Previously, R-loops have been linked to chromatin condensation through the epigenetic modification of phosphorylation at serine 10 residue of histone 3 (H3S10p) [151]. An unexpected link is proposed between R-loops, histone modification and chromatin condensation in which R-loops might trigger the formation of highly compact chromatin. This is either by promoting phosphorylation of Serine 10 residue of histone H3 or by inhibiting its dephosphorylation. It has been suggested that such condensation could spread throughout the genome, leading to enhanced R-T collisions and gene silencing associated with genome instability [151]. Therefore, pathological R-loop formation seems to be associated with H3S10p and chromatin condensation, an association that is in opposition to the link between R-loops and active, hyper-accessible chromatin under physiological conditions [39]. Further, histone modification H3S10p is unique because of its involvement in two opposing processes: transcription activation and chromatin condensation during the cell cycle, as this epigenetic mark increases on mitotic chromosomes instead of being erased upon entry to mitosis [152]. Therefore, a possibility might be that H3S10p, rather than having a direct action on chromatin status, merely acts as a platform to recruit chromatin modifiers and remodelers. There are several nuclear kinases involved in the phosphorylation of H3S10p and this epigenetic mark has been demonstrated to inhibit the histone lysine methyltransferase (KMT) activity of Su(var)3-9 family members such as SUV39H1 and EHMT2/G9a [153]. EHMT2/G9a and EHMT1/G9a-like protein (GLP), amongst other histone methyltransferases, deposit the H3K9me2 mark on euchromatic regions in mammals, while SUV39H1 is associated with the maintenance of constitutive heterochromatin regions [154].
Figure 3. Summary of chromatin modifications associated with the R-loops. R-loops accumulate at the promoter region of genes and are usually associated with an open chromatin structure characterized by H3 acetylation marks, such as H3K27ac and other permissive epigenetic modifications, allowing gene expression and transcription initiation. At terminators, R-loops are formed to induce transcription termination at sites with paused RNAPII, and these R-loops induce the activity of EHMT2/G9a methyltransferase to compact the chromatin by depositing the heterochromatin H3K9me2 mark. Moreover, R-loops are also associated with a closed chromatin architecture that induces repressive marks, such as H3S10p, that might act as a platform for chromatin modifiers and remodelers to alter the chromatin landscape. The antagonism between H3S10p and H3K9me2 prevents the spread of heterochromatin regions across the genome.
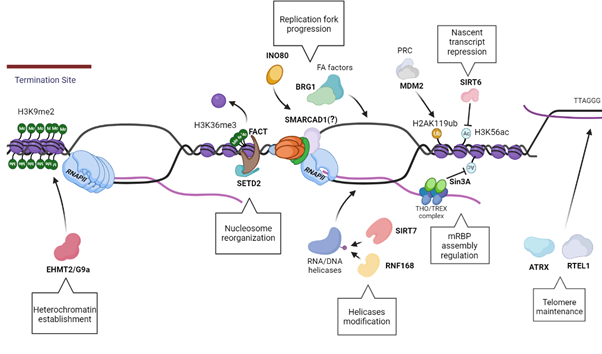
In particular, the ankyrin repeats of G9a and GLP are inhibited by H3S10p, creating an antagonism between H3S10p and H3K9me2 that has been called the “phospho-methyl switch” [155]. The interplay between H3S10p and H3K9me2 has been documented in Drosophila melanogaster, where in the presence of the JIL-1 hypomorph, the main H3S10p kinase, there is a wide-spread loss of euchromatic interbands with propagation of H3K9me2 heterochromatin bands, reinforcing the concept that H3S10p delimits the boundaries of euchromatin in flies by antagonizing heterochromatin propagation [156,157]. These findings have been recently demonstrated also in Mouse Embryonic Fibroblasts (MEFs) and Mouse embryonic stem cells (ESCs) where this antagonism highlights the role of H3S10p in inhibiting the deposition of heterochromatin H3K9me2 mark in mammals [153]. Moreover, transient formation of H3K9me2 has also been shown to accumulate at the terminator regions of genes at RNAPII pausing sites. At the terminator regions, R-loops induce antisense transcription which leads to the formation of dsRNA. This leads to the recruitment of RNAi pathway factors along with lysine methyltransferase EHMT2/G9a, that mediate the formation of repressive epigenetic mark H3K9me2, further reinforcing the pause of RNAPII [150]. This illustrates a direct link between R-loop activity and chromatin architecture modification in which additional chromatin remodelers and modifiers are implicated. Among the many remodelers and modifiers that are thought to have a role in R-loop homeostasis, we highlight a few relevant ones in Table 1.
|
Chro-Mates |
Homologues |
Activity |
Role in R-Loop Processing |
Ref. |
|
EHMT2/G9a |
DmeI |
Modifier |
Methylation of H3K9me2 at heterochromatin regions |
[150,153,158] |
|
SIRT6 |
Sirt6 (Mus musculus) Hst3/4 (Saccharomyces cerevisiae) |
Modifier |
Deacetylation of H3K56ac |
[159,160] |
|
SIRT7 |
Sirt7 (Mus musculus) |
Modifier |
Deacetylation of DDX21 Histone desuccinylation |
[110,161,162] |
|
Sin3A |
Sin3 (Saccharomyces cerevisiae) |
Modifier |
Histone deacetylation through interaction with THO complex |
[163] |
|
RNF168 |
Rnf168 (Mus musculus) |
Modifier |
Ubiquitination of DHX9 |
[164] |
|
MDM2 |
Mdm2 (Mus musculus) |
Remodeler |
Ubiquitination of H2AK119 and genome expression |
[165] |
|
ATRX |
Atrx (Mus musculus) |
Remodeler |
Antagonization of TERRA RNA at telomeric R-loops |
[166,167] |
|
FACT/SETD2 |
yFACT/Set2 (Saccharomyces cerevisiae) |
Remodeler |
Nucleosome reassembly after RNAPII passage |
[168,169] |
|
INO80 |
Ino80 (Saccharomyces cerevisiae) |
Remodeler |
Chromatin relaxation |
[170,171] |
|
BRG1 |
Swi (Saccharomyces cerevisiae) |
Remodeler |
Regulation of chromatin accessibility |
[172] |
|
Fft3/SMARCAD1 |
Fun30 (Saccharomyces cerevisiae) Fft3 |
Remodeler |
Nucleosome turn-over Active fork protection |
[173–176] |
Table 1. Representative Chro-Mates implicated in the processing of R-loops
Figure 4. Schematics showing multiple distinct pathways of R-loop resolution mediated by the Chro-Mates, the chromatin modifying and remodeling factors, described in Section 8 and 9. EHMT2/G9a regulates the preservation of the heterochromatin, especially at the termination sites, in order to permit a successful transcription termination. In addition, the nucleosome reorganization upon RNAPII passage is a vital pathway in R-loops resolution, driven by FACT in concert with SETD2. The modification of RNA/DNA helicases has a role in their recruitment and processivity, such as RNF168 ubiquitinates DHX9, while SIRT7 deacetylates DDX21. The homeostasis of the nascent transcript is also essential to regulate R-loop levels, as seen with Sin3A and SIRT6, histone deacetylases that have a role in creating an optimal chromatin status for a proper transcript export and maturation, along with fork progression. Moreover, the integrity of telomeres is preserved via ATRX and RTEL1, which promotes telomere stability and R-loops resolution. Finally, one of the most critical aspects of R-loop resolution is maintenance of fork stability and replication fork progression. This may require factors involved in resolution of R-loops: as seen with MDM2, that promotes fork progression and gene expression; INO80 possibly resolve R-loops by inducing chromatin relaxation; BRG1 works in concert with FA factors to create an open chromatin structure for the recruitment of R-loops resolvases; and Fft3, which can resolve R-loops in fission yeast with its ortholog SMARCAD1 traveling with the replication forks.
5. Concluding Remarks
Since their discovery, R-loop structures have garnered interest considering the conserved duality of their role in both replication and transcription contexts. In particular, upon conflict between replication and transcription, an inevitable event further accentuated in difficult-to-replicate regions, R-loops pose as one of the major drivers of genome instability [43,49,51,175]. Replication and transcription require the continuous unwinding and rebuilding of nucleosomes and, in this context, chromatin remodelers and modifiers play a central role. Most of the studies to date have focused on the characterization of prevention and removal pathways of RNA:DNA hybrids, while only in recent years has the role of the chromatin landscape been explored. As described above, there are many factors among chromatin remodelers or chromatin modifiers that have a role in R-loop biology. An interesting and contrasting outcome that emerges from this review is that both chromatin accessibility and chromatin condensation are linked to the presence of RNA:DNA hybrids [23,39,150,151]. Given the present state of knowledge, it appears safe to propose a model in which genome stability is ruled by the maintenance of both R-loop and chromatin accessibility homeostasis, depending upon the distinct territory of a genomic region [23,38,94]. An intriguing model would envision the open chromatin as being a hotspot for the formation of R-loops and subsequently, chromatin compaction playing a role in the accumulation and stabilization of these structures, highlighting the close relationship between RNA:DNA hybrids and chromatin landscape. Furthermore, the importance of a fine regulation of R-loop levels is essential to genome integrity, as not only the scheduled but also the unscheduled, yet transient, RNA:DNA hybrids potentially play a role in physiological processes protecting genome integrity. Examples of the latter include the transient RNA:DNA hybrids at the site of DSBs, which are responsible for early stages of DNA repair, efficiently recruiting DNA repair factors. The existence of these structures suggests the interesting possibility that even the unscheduled RNA:DNA hybrids formation must be preserved, as long as they are not persistent and can be timely removed [35,49,142,143,242].
An intriguing unknown mechanism by which various chromatin remodeling and histone modifying factors, associated with distinct state of replication forks and involved in R-loop metabolism, distinguish between the persistent/detrimental RNA:DNA hybrids existing ahead of the fork as an impediment, from the ones associated with the replisome in the form of Okazaki fragments. Furthermore, there might be distinct resolving activities associated with unperturbed and stressed forks, such that the factors associated with unperturbed forks promote fork progression by remodeling the chromatin ahead of the fork, preventing conflicts from occurring, while the resolution of R-loops ahead of stalled fork may involve a distinct mechanism/factor involved in the removal of the conflict and thereby, promote the restart of the fork. It is therefore crucial to determine if there are distinct chromatin features associated with the resolution of harmful, persistent R-loops and transiently-formed, beneficial R-loops. Furthermore, how and which chromatin remodeling/modifying factors, in concert with distinct resolvases, differentiate between these two kinds, from their recognition to their processing, to the maintenance of fine R-loop homeostasis, is yet to be fully elucidated. A recognition of two such distinct regulatory systems with the identification of their components would open up new avenues of research in which chromatin remodeling and modifying activities as a whole will be studied with a new perspective. Targeting molecular pathways and factors that promote the activity of beneficial R-loops while inhibiting the activity of harmful R-loops offers a promising new avenue for therapeutic intervention in a wide array of R-loop-associated diseases.
References
- Robberson, D.L.; Kasamatsu, H.; Vinograd, J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl. Acad. Sci. USA 1972, 69, 737–741.
- Gnatt, A.L.; Cramer, P.; Fu, J.; Bushnell, D.A.; Kornberg, R.D. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science 2001, 292, 1876–1882.
- Bushnell, D.A.; Westover, K.D.; Davis, R.E.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science 2004, 303, 983–988.
- Roy, D.; Lieber, M.R. G Clustering Is Important for the Initiation of Transcription-Induced R-Loops In Vitro, whereas High G Density without Clustering Is Sufficient Thereafter. Mol. Cell Biol. 2009, 29, 3124–3133.
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396.
- Aguilera, A.; García-Muse, T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell 2012, 46, 115–124.
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618.
- Allison, D.F.; Wang, G.G. R-loops: Formation, function, and relevance to cell stress. Cell Stress 2019, 3, 38–46.
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411.
- Chien, Y.H.; Davidson, N. Rna-DNA Hybrids Are More Stable Than DNA-DNA Duplexes in Concentrated Perchlorate and Trichloroacetate Solutions. Nucleic Acids Res. 1978, 5, 1627–1637.
- Sugimoto, N.; Nakano, S.; Katoh, M.; Matsumura, A.; Nakamuta, H.; Ohmichi, T.; Yoneyama, M.; Sasaki, M. Thermodynamic Parameters to Predict Stability of Rna/DNA Hybrid Duplexes. Biochemistry 1995, 34, 11211–11216.
- Wongsurawat, T.; Jenjaroenpun, P.; Kwoh, C.K.; Kuznetsov, V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012, 40, e16.
- Lang, K.S.; Hall, A.N.; Merrikh, C.N.; Ragheb, M.; Tabakh, H.; Pollock, A.J.; Woodward, J.J.; Dreifus, J.E.; Merrikh, H. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 2017, 170, 787–799.e18.
- Leela, J.K.; Syeda, A.H.; Anupama, K.; Gowrishankar, J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 258–263.
- Wahba, L.; Costantino, L.; Tan, F.J.; Zimmer, A.; Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016, 30, 1327–1338.
- Huertas, P.; Aguilera, A. Cotranscriptionally Formed DNA: RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol. Cell 2003, 12, 711–721.
- Xu, W.; Xu, H.; Li, K.; Fan, Y.X.; Liu, Y.; Yang, X.R.; Sun, Q.W. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 2017, 3, 704–714.
- Xu, W.; Li, K.; Li, S.; Hou, Q.C.; Zhang, Y.S.; Liu, K.P.; Sun, Q.W. The R-Loop Atlas of Arabidopsis Development and Responses to Environmental Stimuli. Plant Cell 2020, 32, 888–903.
- Ginno, P.A.; Lim, Y.W.; Lott, P.L.; Korf, I.; Chédin, F. GC skew at the 59 and 39 ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013, 23, 1590–1600.
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.Q.; Chedin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178.
- Chedin, F.; Benham, C.J.; Musier-Forsyth, K. Emerging roles for R-loop structures in the management of topological stress. J. Biol. Chem. 2020, 295, 4684–4695.
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505.
- Lockhart, A.; Pires, V.B.; Bento, F.; Kellner, V.; Luke-Glaser, S.; Yakoub, G.; Ulrich, H.D.; Luke, B. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep. 2019, 29, 2890–2900.e5.