
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emiliano Fabiani | + 2121 word(s) | 2121 | 2021-08-26 09:37:31 | | | |
| 2 | Vivi Li | Meta information modification | 2121 | 2021-08-30 04:55:50 | | | | |
| 3 | Conner Chen | Meta information modification | 2121 | 2021-09-22 04:13:45 | | |
Video Upload Options
Forkhead box (FOX) proteins are a group of transcriptional factors implicated in different cellular functions such as differentiation, proliferation and senescence. A growing number of studies have focused on the relationship between FOX proteins and cancers, particularly hematological neoplasms such as acute myeloid leukemia (AML). FOX proteins are widely involved in AML biology, including leukemogenesis, relapse and drug sensitivity. Here we explore the role of FOX transcription factors in the major AML entities, according to “The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia”, and in the context of the most recurrent gene mutations identified in this heterogeneous disease.
1. Introduction
2. Forkhead Box Proteins in RUNX1-RUNX1T1 Acute Myeloid Leukemia
| Recurrent Abnormalities in Acute Myeloid Leukemia | FOX Family Member | Biological Process | References |
|---|---|---|---|
| t(8;21); RUNX1-RUNX1T1 | FOXO1 | Self-renewal and Differentiation | Lin et al. [19] |
| PML-RARA | FOXO3A FOXC1 |
Apoptosis and Granulocytic Differentiation Granulocytic Differentiation and Epigenetic Regulation |
Sakoe et al. [35] Somerville et al. [36] and Fabiani et al. [37] |
| NPM1 | FOXM1 | Cell Proliferation, Division and Chemoresistance | Laoukili et al. [38] Nakamura et al. [39] Khan et al. [40] |
| FLT3 ITD | FOXO3A FOXO1 FOXM1 |
Apoptosis, Survival and Proliferation Cell growth, Apoptosis and antioxidant defences Survival, Apoptosis and Chemoresistance |
Scheijen et al. [41] Seedhouse et al. [42] Liu et al. [43] |
| IDH 1-2 | FOXOs | Cellular Differentiation and Tumor Suppression/Progression and Epigenetic Instability | Charitou et al. [44] |
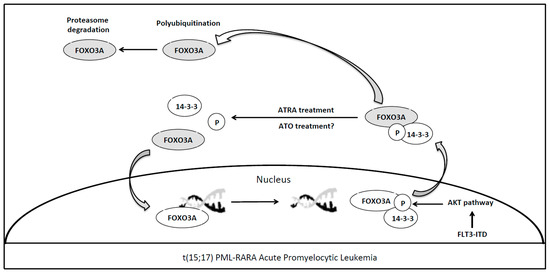
3. Forkhead Box Proteins in PML/RARA (Promyelocytic Leukemia/Retinoic Acid Receptor-Alpha) Acute Promyelocytic Leukemia
References
- Weigel, D.; Jürgens, G.; Küttner, F.; Seifert, E.; Jäckle, H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 1989, 57, 645–658.
- Hannenhalli, S.; Kaestner, K.H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009, 10, 233–240.
- Jackson, B.C.; Carpenter, C.; Nebert, D.W.; Vasiliou, V. Update of human and mouse forkhead box (FOX) gene families. Hum. Genom. 2010, 4, 345–352.
- Wang, J.; Li, W.; Zhao, K.; Fu, W.; Zheng, X.; Pang, X.; Du, G. Members of FOX family could be drug targets of cancers. Pharmacol. Ther. 2018, 181, 183–196.
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570.
- He, L.; Gomes, A.P.; Wang, X.; Yoon, S.O.; Lee, G.; Nagiec, M.J.; Cho, S.; Chavez, A.; Islam, T.; Yu, Y.; et al. mTORC1 Promotes Metabolic Reprogramming by the Suppression of GSK3-Dependent Foxk1 Phosphorylation. Mol. Cell 2018, 70, 949–960.
- Maachani, U.B.; Shankavaram, U.; Kramp, T.; Tofilon, P.; Camphausen, K.; Tandle, A.T. FOXM1 and STAT3 interaction confers radioresistance in glioblastoma cells. Oncotarget 2016, 7, 77365–77377.
- Alvarez-Fernández, M.; Medema, R.H. Novel functions of FoxM1: From molecular mechanisms to cancer therapy. Front. Oncol. 2013, 3, 30.
- Ackermann, S.; Kocak, H.; Hero, B.; Ehemann, V.; Kahlert, Y.; Oberthuer, A.; Roels, F.; Theißen, J.; Odenthal, M.; Berthold, F.; et al. FOXP1 inhibits cell growth and attenuates tumorigenicity of neuroblastoma. BMC Cancer 2014, 14, 840.
- Wang, P.; Lv, C.; Zhang, T.; Liu, J.; Yang, J.; Guan, F.; Hong, T. FOXQ1 regulates senescence-associated inflammation via activation of SIRT1 expression. Cell Death Dis. 2017, 8, e2946.
- Macedo, J.C.; Vaz, S.; Bakker, B.; Ribeiro, R.; Bakker, P.L.; Escandell, J.M.; Ferreira, M.G.; Medema, R.; Foijer, F.; Logarinho, E. FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nat. Commun. 2018, 9, 2834.
- Song, B.N.; Chu, I.S. A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp. Mol. Med. 2018, 50, e418.
- Gao, Y.F.; Zhu, T.; Mao, X.Y.; Mao, C.X.; Li, L.; Yin, J.Y.; Zhou, H.H.; Liu, Z.Q. Silencing of Forkhead box D1 inhibits proliferation and migration in glioma cells. Oncol. Rep. 2017, 37, 1196–1202.
- Lam, E.W.; Brosens, J.J.; Gomes, A.R.; Koo, C.Y. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat. Rev. Cancer 2013, 13, 482–495.
- Karadedou, C.T.; Gomes, A.R.; Chen, J.; Petkovic, M.; Ho, K.K.; Zwolinska, A.K.; Feltes, A.; Wong, S.Y.; Chan, K.Y.; Cheung, Y.N.; et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene 2012, 31, 1845–1858.
- Rocca, D.L.; Wilkinson, K.A.; Henley, J.M. SUMOylation of FOXP1 regulates transcriptional repression via CtBP1 to drive dendritic morphogenesis. Sci. Rep. 2017, 7, 877.
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859.
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405.
- Lin, S.; Ptasinska, A.; Chen, X.; Shrestha, M.; Assi, S.A.; Chin, P.S.; Imperato, M.R.; Aronow, B.J.; Zhang, J.; Weirauch, M.T.; et al. A FOXO1-induced oncogenic network defines the AML1-ETO preleukemic program. Blood 2017, 130, 1213–1222.
- Müller, A.M.; Duque, J.; Shizuru, J.A.; Lübbert, M. Complementing mutations in core binding factor leukemias: From mouse models to clinical applications. Oncogene 2008, 27, 5759–5773.
- Tonks, A.; Pearn, L.; Musson, M.; Gilkes, A.; Mills, K.I.; Burnett, A.K.; Darley, R.L. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia 2007, 21, 2495–2505.
- Jagani, Z.; Singh, A.; Khosravi-Far, R. FoxO tumor suppressors and BCR-ABL-induced leukemia: A matter of evasion of apoptosis. Biochim. Biophys. Acta 2008, 1785, 63–84.
- Kornblau, S.M.; Singh, N.; Qiu, Y.; Chen, W.; Zhang, N.; Coombes, K.R. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin. Cancer Res. 2010, 16, 1865–1874.
- Obrador-Hevia, A.; Serra-Sitjar, M.; Rodríguez, J.; Villalonga, P.; Fernández de Mattos, S. The tumour suppressor FOXO3 is a key regulator of mantle cell lymphoma proliferation and survival. Br. J. Haematol. 2012, 156, 334–345.
- Xie, L.; Ushmorov, A.; Leithäuser, F.; Guan, H.; Steidl, C.; Färbinger, J.; Pelzer, C.; Vogel, M.J.; Maier, H.J.; Gascoyne, R.D.; et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood 2012, 119, 3503–3511.
- Yao, S.; Mahmud, Z.; Sachini, N.; Aimjongjun, S.; Saavedra-García, P.; Lam, E.W. Characterization of FOXO Acetylation. Methods Mol. Biol. 2019, 1890, 77–90.
- Rehman, A.; Kim, Y.; Kim, H.; Sim, J.; Ahn, H.; Chung, M.S.; Shin, S.J.; Jang, K. FOXO3a expression is associated with lymph node metastasis and poor disease-free survival in triple-negative breast cancer. J. Clin. Pathol. 2018, 71, 806–813.
- Hou, T.; Li, Z.; Zhao, Y.; Zhu, W.G. Mechanisms controlling the anti-neoplastic functions of FoxO proteins. Semin. Cancer Biol. 2018, 50, 101–114.
- Pan, C.W.; Jin, X.; Zhao, Y.; Pan, Y.; Yang, J.; Karnes, R.J.; Zhang, J.; Wang, L.; Huang, H. AKT-phosphorylated FOXO1 suppresses ERK activation and chemoresistance by disrupting IQGAP1-MAPK interaction. EMBO J. 2017, 36, 995–1010.
- Zhu, H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia. Oncol. Rep. 2014, 32, 1327–1334.
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827.
- Calnan, D.R.; Brunet, A. The FoxO code. Oncogene 2008, 27, 2276–2288.
- Hui, R.C.; Francis, R.E.; Guest, S.K.; Costa, J.R.; Gomes, A.R.; Myatt, S.S.; Brosens, J.J.; Lam, E.W. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol. Cancer Ther. 2008, 7, 670–678.
- Han, C.Y.; Cho, K.B.; Choi, H.S.; Han, H.K.; Kang, K.W. Role of FoxO1 activation in MDR1 expression in adriamycin-resistant breast cancer cells. Carcinogenesis 2008, 29, 1837–1844.
- Sakoe, Y.; Sakoe, K.; Kirito, K.; Ozawa, K.; Komatsu, N. FOXO3A as a key molecule for all-trans retinoic acid–induced granulocytic differentiation and apoptosis in acute promyelocytic leukemia. Blood 2010, 115, 3787–3795.
- Somerville, T.D.; Wiseman, D.H.; Spencer, G.J.; Huang, X.; Lynch, J.T.; Leong, H.S.; Williams, E.L.; Cheesman, E.; Somervaille, T.C. Frequent Derepression of the Mesenchymal Transcription Factor Gene FOXC1 in Acute Myeloid Leukemia. Cancer Cell 2015, 28, 329–342.
- Fabiani, E.; Falconi, G.; Noguera, N.I.; Saulle, E.; Cicconi, L.; Divona, M.; Banella, C.; Picardi, A.; Cerio, AM.; Boe, L.; et al. The forkhead box C1 (FOXC1) transcription factor is downregulated in acute promyelocytic leukemia. Oncotarget 2017, 8, 84074–84085.
- Laoukili, J.; Stahl, M.; Medema, R.H. FoxM1: At the crossroads of ageing and cancer. Biochim. Biophys. Acta 2007, 1775, 92–102.
- Nakamura, S.; Hirano, I.; Okinaka, K.; Takemura, T.; Yokota, D.; Ono, T.; Shigeno, K.; Shibata, K.; Fujisawa, S.; Ohnishi, K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis 2010, 31, 2012–2021.
- Khan, I.; Halasi, M.; Patel, A.; Schultz, R.; Kalakota, N.; Chen, Y.H.; Aardsma, N.; Liu, L.; Crispino, J.D.; Mahmud, N.; et al. FOXM1 contributes to treatment failure in acute myeloid leukemia. JCI Insight 2018, 3.
- Scheijen, B.; Ngo, H.T.; Kang, H.; Griffin, J.D. FLT3 receptors with internal tandem duplications promote cell viability and proliferation by signaling through Foxo proteins. Oncogene 2004, 23, 3338–3349.
- Seedhouse, C.H.; Mills, K.I.; Ahluwalia, S.; Grundy, M.; Shang, S.; Burnett, A.K.; Russell, N.H.; Pallis, M. Distinct poor prognostic subgroups of acute myeloid leukaemia, FLT3-ITD and P-glycoprotein-positive, have contrasting levels of FOXO1. Leuk. Res. 2014, 38, 131–137.
- Liu, L.L.; Zhang, D.H.; Mao, X.; Zhang, X.H.; Zhang, B. Over-expression of FoxM1 is associated with adverse prognosis and FLT3-ITD in acute myeloid leukemia. Biochem. Biophys. Res. Commun. 2014, 446, 280–285.
- Charitou, P.; Rodriguez-Colman, M.; Gerrits, J.; van Triest, M.; Groot Koerkamp, M.; Hornsveld, M.; Holstege, F.; Verhoeven-Duif, N.M.; Burgering, B.M. FOXOs support the metabolic requirements of normal and tumor cells by promoting IDH1 expression. EMBO Rep. 2015, 16, 456–466.
- Grignani, F.; Ferrucci, P.F.; Testa, U.; Talamo, G.; Fagioli, M.; Alcalay, M.; Mencarelli, A.; Grignani, F.; Peschle, C.; Nicoletti, I.; et al. The acute promyelocytic leukemia-specific PML-RAR alpha fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 1993, 74, 423–431.
- Altucci, L.; Rossin, A.; Raffelsberger, W.; Reitmair, A.; Chomienne, C.; Gronemeyer, H. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat. Med. 2001, 7, 680–686.
- Birkenkamp, K.U.; Coffer, P.J. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem. Soc. Trans. 2003, 31, 292–297.
- Komatsu, N.; Watanabe, T.; Uchida, M.; Mori, M.; Kirito, K.; Kikuchi, S.; Liu, Q.; Tauchi, T.; Miyazawa, K.; Endo, H.; et al. A member of Forkhead transcription factor FKHRL1 is a downstream effector of STI571-induced cell cycle arrest in BCR-ABL-expressing cells. J. Biol. Chem. 2003, 278, 6411–6419.
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S.; Ferrara, F.; Fazi, P.; Cicconi, L.; Di Bona, E.; et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013, 369, 111–121.
- Zhang, L.; Liu, L.; Zhan, S.; Chen, L.; Wang, Y.; Zhang, Y.; Du, J.; Wu, Y.; Gu, L. Arsenic Trioxide Suppressed Migration and Angiogenesis by Targeting FOXO3a in Gastric Cancer Cells. Int. J. Mol. Sci. 2018, 19.
- Alfonso, V.; Iaccarino, L.; Ottone, T.; Cicconi, L.; Lavorgna, S.; Divona, M.; Cairoli, R.; Cristiano, A.; Ciardi, C.; Travaglini, S.; et al. Early and sensitive detection of PML-A216V mutation by droplet digital PCR in ATO-resistant acute promyelocytic leukemia. Leukemia 2019.
- Soncini, M.; Santoro, F.; Gutierrez, A.; Frigè, G.; Romanenghi, M.; Botrugno, O.A.; Pallavicini, I.; Pelicci, P.; Di Croce, L.; Minucci, S. The DNA demethylating agent decitabine activates the TRAIL pathway and induces apoptosis in acute myeloid leukemia. Biochim. Biophys. Acta 2013, 1832, 114–120.




