Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jin, B. Jasmonic Acid in Plant Responses. Encyclopedia. Available online: https://encyclopedia.pub/entry/13260 (accessed on 07 February 2026).
Jin B. Jasmonic Acid in Plant Responses. Encyclopedia. Available at: https://encyclopedia.pub/entry/13260. Accessed February 07, 2026.
Jin, Biao. "Jasmonic Acid in Plant Responses" Encyclopedia, https://encyclopedia.pub/entry/13260 (accessed February 07, 2026).
Jin, B. (2021, August 17). Jasmonic Acid in Plant Responses. In Encyclopedia. https://encyclopedia.pub/entry/13260
Jin, Biao. "Jasmonic Acid in Plant Responses." Encyclopedia. Web. 17 August, 2021.
Copy Citation
As sessile organisms, plants must tolerate various environmental stresses. Plant hormones play vital roles in plant responses to biotic and abiotic stresses. Among these hormones, jasmonic acid (JA) and its precursors and derivatives (jasmonates, JAs) play important roles in the mediation of plant responses and defenses to biotic and abiotic stresses and have received extensive research attention.
abiotic stress
biotic stress
crosstalk
defense response
jasmonic acid
plant hormones
1. Introduction
Plants live in the ever-changing natural environment and encounter many factors that are not suitable for growth or even survival [1]. These factors are generally divided into biotic and abiotic stressors. The main biotic stressors include pathogenic bacterial diseases, insects, and herbivores, while drought, saline or alkaline conditions, and extreme temperature are considered abiotic stressors [2][3]. As sessile organisms, plants have evolved sophisticated response mechanisms to resist, mitigate, or recover from these stressors. In this respect, the fundamental and important biological question is how plants perceive stress signals and respond to various adverse environmental conditions. Over the past two decades, plant hormones, which are vital regulators involved in sensing and transmitting various environmental signals and subsequent defense responses, have received intense research attention. Currently, nine major classes of natural plant hormones have been described that underlie numerous reactions to environmental signals, including auxin, cytokinin, gibberellin (GA), abscisic acid (ABA), ethylene (ET), brassinosteroid (BR), jasmonic acid (JA), salicylic acid (SA), and strigolactone [4]. Among these hormones, JA is ubiquitous in higher plant species and therefore has attracted great attention in the field of plant stress response and defense mechanisms [5].
JA and its derivatives, including its methyl ester (MeJA) and its isoleucine conjugate (JA-Ile), are collectively called jasmonates (JAs) [6]. Aromatic MeJA was first isolated from the essential oil of Jasminum grandiflorum L. [7]. Free JA was examined and isolated from the culture filtrate of Lasiodiplodia theobromae, and was found to inhibit plant growth [8], providing the first report of the physiological function of JA. JAs play a variety of regulatory roles in plant growth and development, for example, axis elongation during embryogenesis, flower development, leaf senescence, root formation, and stomatal opening [9][10]. In addition to growth and development, many studies have shown that JAs improve plant stress tolerance via JA signaling pathways under various adverse environmental conditions.
Some molecular models of plant–environment interactions include microbe-associated molecular patterns (MAMPs), herbivore-associated molecular patterns (HAMPs), and damage-associated molecular patterns (DAMPs), which are mainly derived from attacking organisms, cell damage, and abiotic stresses (e.g., salt, drought, heavy metals, cold, etc.) (Figure 1) [11][12]. In general, these molecular patterns are associated with JA signaling pathways. The receptor-active conjugated complex JA-Ile is a core component of JA signaling pathways [13][14][15]. As inducing signals, environmental stimuli are recognized by cell surface receptors, triggering de novo synthesis of JA-Ile from plastid lipids, which ultimately results in downstream transcription factor interactions and promotion of growth, development, and specific protective mechanisms in plants [12]. In this reaction, JA-Ile acts to facilitate the interaction between jasmonate zinc-finger inflorescence meristem (JAZ) and coronatine insensitive 1 (COI1) protein within the Skp1p–cullin–F-box protein (SCF) complex, and promotes degradation of JAZ proteins (JAZx, JAZy, and JAZz), resulting in the activation of JAZ-interacting transcription factors (TFs: TFa, TFb, TFc) (Figure 1). These TFs regulate the expression of numerous genes in response to both biotic (e.g., herbivores and fungi) and abiotic (e.g., salt, drought, heavy metals, and cold) stresses and promote specific protection mechanisms (Figure 1) [16][17][18][19]. Therefore, JAs are functionally significant and have attracted extensive research interest in the field of plant–environment interactions.
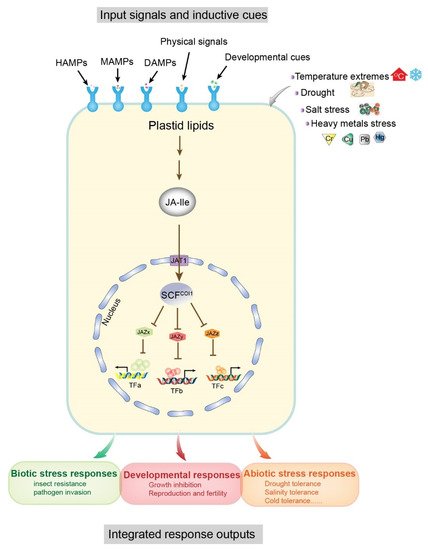
Figure 1. Input signals and output responses of the core jasmonoyl-L-isoleucine (JA-Ile) signaling pathway. Associations of the modular design of JA-Ile signaling with a variety of inductive cues (top) and physiological output responses (bottom) are shown as a schematic diagram. Inductive signals include biotic stresses (microbe and herbivore) and damage related to molecular patterns (MAMPs, microbe-associated; HAMPs, herbivore-associated; and DAMPs, damage-associated) arising from damaged plant cells, attacking organisms, and abiotic stresses. Pattern recognition receptors on the cell surface recognize the signals and resynthesize JA-Ile from plastid lipids. The degradation of multiple JAZ proteins (exemplified by JAZx-z) is promoted by the activation of the E3 ubiquitin ligase SCFCOI1 and the 26S proteasome. Subsequently, JAZ-interacting transcription factors (exemplified by TFa–c), which govern diverse physiological output responses involved in growth, development, and tolerance to biotic and abiotic stresses, are upregulated due to the degradation of JAZ. The conserved core pathway may be repurposed to connect other input signals and transcription factors. Abbreviations: JA-Ile, jasmonoyl-L-isoleucine; MAMPs, microbe-associated molecular patterns; HAMPs, herbivore-associated molecular patterns; DAMPs, damage-associated molecular patterns; JAZ, jasmonate ZIM domain; SCF, [kinetochore protein 1 (SKP1)-CULLIN1 (CUL1)-F-box]; COI1, coronatine insensitive1; TF, transcription factor. Modified from [12].
2. Biosynthesis and Metabolism of JAs
2.1. JA Biosynthesis
Several reviews of the JA biosynthesis pathway have been published previously. These reviews provide important information on the biosynthesis reactions, genes, and enzymes in this pathway, including explanations of the mechanisms determining enzymatic crystal structure and substrate specificity, and ultimately describe the regulation of JA biosynthesis [20][21][22][23]. Recently, several membrane transporters, including JASSY, comatose (CTS), and jasmonate transporter 1 (JAT1), were reported to function in the JA biosynthesis pathway. In Figure 2, we provide an updated summary of the JA synthesis process, including the reaction steps and names of enzymes and substrates.
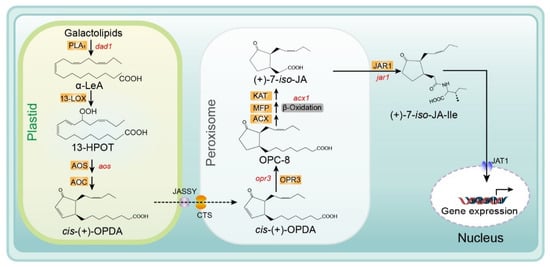
Figure 2. Biosynthesis of JA/JA-Ile initiated by the release of α-linolenic acid from galactolipid. De novo synthesis of JA occurs in the plastids, peroxisomes, and cytoplasm, and JA-Ile is then transported to the nucleus for gene expression. Major genes are indicated in red and enzyme names are highlighted in orange. Abbreviations for compounds: α-LeA, α-linolenic acid; 13-HPOT, (13S)-hydroperoxyoctadecatrienoic acid; cis- (+)-OPDA, cis- (+)-12-oxophytodienoic acid; OPC-8, 3-oxo-2- (2-pentenyl)-cyclopentane-1-octanoic acid; (+)-7-iso-JA, jasmonic acid; (+)-7-iso-JA-Ile, jasmonic acid isoleucine conjugate. Abbreviations for enzymes/proteins: PLA1, phospholipase A1; 13-LOX, 13-lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR3, OPDA reductase3; ACX, acyl-CoA oxidase; MFP, multifunctional protein; KAT, 3-ketoacyl-CoA thiolase; JAR1, JA-amino acid synthetase; JASSY, a chloroplast outer membrane protein; CTS, an ABC transporter of the peroxisomal membrane; JAT1, jasmonate transporter 1 (Redrawn based on [24]).
The substrate for the biosynthesis of JA is α-linolenic acid (18:3) (α-LeA), which is released from the sn1 position of galactolipids on chloroplast membranes through the action of phospholipase A1 (PLA1) [25]. Next, the substrate is converted to (9S,13S)-12-oxo-phytodienoic acid (OPDA) through the actions of 13-lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC), respectively. All enzymes and products involved in this pathway are located in plastids (chloroplasts). Subsequently, OPDA is transferred from the chloroplast to the peroxisome and transformed into 3-oxo-2- (cis-2ʹ-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0) by OPDA reductase 3 (OPR3) [6][26][27]. The mechanism of OPDA transport from chloroplasts to the peroxisome remains unclear. Guan et al. identified a protein located on the chloroplast outer envelope membranes that is responsible for exporting OPDA and named it JASSY [28]. For the recipient, CTS—an ABC transporter on the peroxisomal membrane—is the main mediator of OPDA import into peroxisomes [29]. Subsequently, OPC-8 is transformed into (+)-7-iso-JA through three β-oxidation reactions by three different enzymes: acyl-CoA oxidase (ACX), multifunctional protein (MEP), and L-3-ketoacyl CoA thiolase (KAT) (Figure 2.) [9][30]. Next, (+)-7-iso-JA is transported to the cytoplasm, where it is conjugated with isoleucine (Ile) to form (+)-7-iso-JA-Ile—which is considered the most bioactive JA compound—under the action of JAR1 (a JA-Ile synthesizing enzyme). Finally, JA-Ile is transported into the nucleus by the ABC transporter JAT1, where it participates in the subsequent steps of the JA signaling pathway [15] (Figure 2).
2.2. JA Metabolism
After biosynthesis of JA and JA-Ile, JA derivatives including active, inactive, and partially active compounds are derived mainly from JA and JA-Ile through at least 12 metabolic pathways. These metabolic pathways include conjugation with amino acids, decarboxylation, sulfation, hydroxylation, carboxylation, o-glycosylation, methylation, esterification, and lactone formation. Consequently, diverse JA derivatives are formed (Figure 3) [24][31].
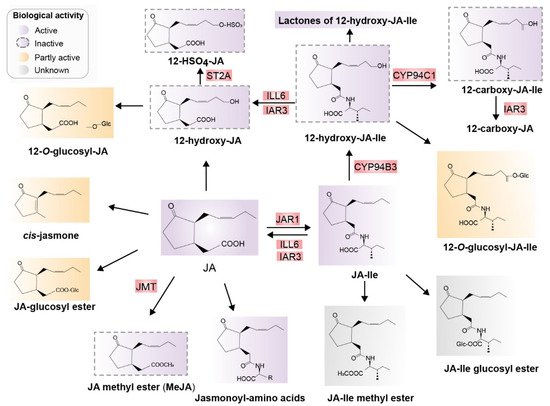
Figure 3. Major jasmonates arising from the metabolic conversion of JA. JA can be methylated to form JA-Me (MeJA) or JA glucosyl ester, decarboxylated to cis-jasmone, or hydroxylated to 12-OH-JA. 12-OH-JA can be sulfated and O-glucosylated, and conjugated with amino acids, preferring isoleucine, which results in JA-Ile. JA-Ile is methylated to JA-Me-Ile. Moreover, 12-hydroxylation of JA-Ile, carboxylation of 12-OH-JA-Ile, O-glucosylation of JA-Ile, and JA-Ile glucosyl ester formation are also possible. Known enzymes involved in these transformations are highlighted in red. Biologically active compounds are in purple and inactive compounds are indicated with dotted outlines in purple. Partially active compounds are shown in yellow, and unknown compounds in gray. Abbreviations: JA, jasmonic acid; JA-Ile, jasmonyl isoleucine; JMT, JA methyltransferase; JAR1, jasmonoyl isoleucine synthetase; CYP94B3, JA-Ile-12-hydroxylase; CYP94C1, 12-OH-JA-Ile carboxylase; the amidohydrolases IAR3 and ILL6; JMT, JA methyltransferase; ST2A, 12-OH-JA sulfotransferase (Redrawn based on [24]).
References
- Vanwallendael, A.; Soltani, A.; Emery, N.C.; Peixoto, M.M.; Olsen, J.; Lowry, D.B. A molecular view of plant local adaptation: Incorporating stress-response networks. Annu. Rev. Plant Biol. 2019, 70, 559–583.
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, F1000.
- Alonso, C.; Ramos-Cruz, D.; Becker, C. The role of plant epigenetics in biotic interactions. New Phytol. 2019, 221, 731–737.
- Kamiya, Y. Plant hormones: Versatile regulators of plant growth and development. Annu. Rev. Plant. Biol. 2010, 61.
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206.
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479.
- Demole, E.; Lederer, E.; Mercier, D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caractéristique de l’essence de jasmin. Helv. Chim. Acta 1962, 45, 675–685.
- Aldridge, D.C.; Galt, S.; Giles, D.; Turner, W.B. Metabolites of Lasiodiplodia theobromae. J. Chem. Soc. C Org. Chem. 1971, 1623–1627.
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359.
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 2019, 165, 90–100.
- Acevedo, F.E.; Rivera-Vega, L.J.; Chung, S.H.; Ray, S.; Felton, G.W. Cues from chewing insects—the intersection of DAMPs, HAMPs, MAMPs and effectors. Curr. Opin. Plant. Biol. 2015, 26, 80–86.
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant. Biol. 2018, 69, 387–415.
- Marquis, V.; Smirnova, E.; Poirier, L.; Zumsteg, J.; Schweizer, F.; Reymond, P.; Heitz, T. Stress- and pathway-specific impacts of impaired jasmonoyl-isoleucine (JA-Ile) catabolism on defense signalling and biotic stress resistance. Plant Cell Environ. 2020, 43, 1558–1570.
- Zander, M. Many ways to repress! JAZ’s agony of choices. Mol. Plant 2021, 14, 714–716.
- Li, Q.; Zheng, J.; Li, S.; Huang, G.; Skilling, S.J.; Wang, L.; Li, L.; Li, M.; Yuan, L.; Liu, P. Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential forjasmonate signaling. Mol. Plant 2017, 10, 695–708.
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058.
- Takeuchi, J.; Fukui, K.; Seto, Y.; Takaoka, Y.; Okamoto, M. Ligand–receptor interactions in plant hormone signaling. Plant J. 2021, 105, 290–306.
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2020, 1–29.
- Ali, M.S.; Baek, K.H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621.
- Jang, G.; Yoon, Y.; Choi, Y. Do Crosstalk with jasmonic acid integrates multiple responses in plant development. Int. J. Mol. Sci. 2020, 21, 305.
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth–defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81.
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis—Structure, function, regulation. Phytochemistry 2009, 70, 1532–1538.
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156.
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321.
- Scherer, G.F.E.; Ryu, S.B.; Wang, X.; Matos, A.R.; Heitz, T. Patatin-related phospholipase A: Nomenclature, subfamilies and functions in plants. Trends Plant Sci. 2010, 15, 693–700.
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048.
- Han, G.Z. Evolution of jasmonate biosynthesis and signalling mechanisms. J. Exp. Bot. 2017, 68, 1323–1331.
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575.
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005, 137, 835–840.
- Yang, T.-H.; Lenglet-Hilfiker, A.; Stolz, S.; Glauser, G.; Farmer, E.E. Jasmonate precursor biosynthetic enzymes LOX3 and LOX4 control wound-response growth restriction. Plant Physiol. 2020, 184, 1172–1180.
- Ghasemi Pirbalouti, A.; Sajjadi, S.E.; Parang, K. A review (research and patents) on jasmonic acid and its derivatives. Arch. Pharm. 2014, 347, 229–239.
More
Information
Subjects:
Plant Sciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
17 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




