
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Geon Hui Son | + 2582 word(s) | 2582 | 2021-07-16 10:49:35 | | | |
| 2 | Ron Wang | + 1 word(s) | 2583 | 2021-08-09 10:24:30 | | |
Video Upload Options
SUPPRESSOR OF rps4-RLD1 (SRFR1) is known as a negative regulator by forming an immune complex with resistance proteins and transcription factors of the plant immune system in Arabidopsis. Mutations in SRFR1, identified in a suppressor screen, activated EDS1-dependent ETI to Pseudomonas syringae pv. tomato (Pto) DC3000. Besides, mutations in SRFR1 boosted defense responses to the generalist chewing insect Spodoptera exigua and the sugar beet cyst nematode Heterodera schachtii. In the current study, we report that mutations in SRFR1 enhance susceptibility to the fungal necrotrophs Fusarium oxysporum f. sp. lycopersici (FOL) and Botrytis cinerea in Arabidopsis. The slsrfr1 tomato mutants generated by a CRISPR/Cas9 system increased expression of SA-pathway defense genes and enhanced resistance to Pto DC3000. In contrast, slsrfr1 mutants elevate susceptibility to FOL. Together, these data suggest that SRFR1 is functionally conserved in both Arabidopsis and tomato and functions antagonistically as a negative regulator to biotrophic pathogens and a positive regulator to necrotrophic pathogens.
1. Introduction
2. Enhanced Susceptibility to the Necrotrophic Fungal Pathogens in Arabidopsis srfr1 Mutants
Arabidopsis srfr1 mutants were mainly involved in EDS1-dependent ETI responses against Pto DC3000 [1][3][8] and in the resistance to chewing insect S. exigua and the sugar beet cyst nematode H. schachtii, which is an obligate biotrophic pathogen [7]. FOL has been reported to cause vascular wilt disease in Arabidopsis and tomato. In FOL - plant interactions, initial fungal infection occurs primarily in the roots, resulting in disruption of vascular tissues, chlorosis, and necrosis, which leads to plant death [9],[10]. The Arabidopsis wild-type accessions Col-0 and RLD, and srfr1-1 and srfr1-2, two recessive alleles of SRFR1 in the RLD background, were plug-inoculated with FOL. The wild-type RLD was more resistant to FOL than Col-0 (Figure 1a). Lesion size in Col-0 was three times larger than in RLD (Figure 1b), suggesting RLD is naturally resistant to FOL. Interestingly, the srfr1-1 and srfr1-2 mutants displayed enhanced susceptibility to FOL compared to RLD, as reflected by more severe symptoms, such as increased chlorosis (Figure 1a) and increased lesion size (Figure 1b).
B. cinerea is a necrotrophic fungal pathogen that destroys plant cells at the early stage of infection resulting in widespread tissue injury [11][12]. The wild-type RLD and srfr1 mutants were drop-inoculated with B. cinerea. As shown in Figure 1c, the lesion areas were more extensive in srfr1 mutants than that in RLD. The lesion area in srfr1 mutants was twice larger than that in the wild-type (Figure 1d). These results demonstrate that mutations in SRFR1 enhanced susceptibility to FOL and B. cinerea, indicating SRFR1 functions as a positive regulator of plant disease resistance against necrotrophic fungal pathogens.
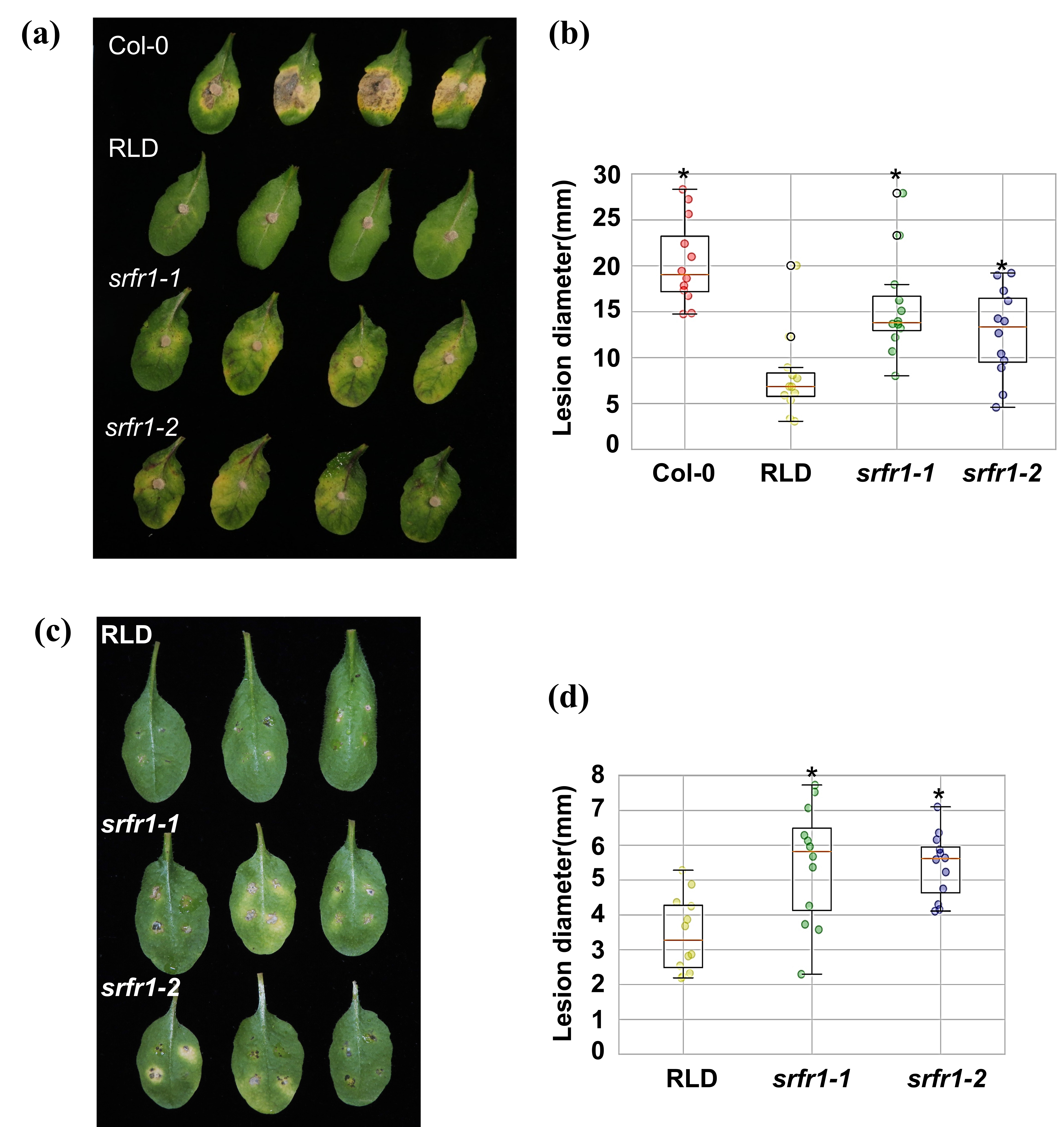
Figure 1. Response of Arabidopsis srfr1 mutants to the necrotrophic fungal pathogens. (a) Infection with conidia and mycelial plugs on Arabidopsis RLD, srfr1-1, and srfr1-2 mutants. Detached leaves from 4-week-old plants were inoculated with 4 mm-diameter plugs of F. oxysproum. Photographs were taken at 14 DPI; (b) Box plots of lesion size at 14 DPI in Col-0, RLD, srfr1-1, and srfr1-2. The y-axis displays the measured diameter of disease lesion (mm, n=12) in each plant. The box ranges were determined from the twenty-fifth to the seventy-fifth percentiles (P<0.05).; (c) Fungal spores of B. cinerea grown on PDA were harvested and inoculated onto detached leaves of Arabidopsis RLD, srfr1-1, and srfr1-2 mutants at a concentration of 1 X 108 spores/ml. Photographs were taken at 5 DPI; (d) Box plots of lesion size at 5 DPI in RLD, srfr1-1, and srfr1-2. The y-axis displays the measured diameter of disease lesions (mm, n=12) in each plant. The box ranges were determined from the twenty-fifth to the seventy-fifth percentiles (P<0.01).
3. Altered Morphology and Expression of Defense Marker Genes in CRISPR/Cas9-Edited slsrfr1 Plants
In Arabidopsis, RLD srfr1-1 and srfr1-2 mutants exhibit normal and wild-type-like morphology, although there is a slight decrease in growth. However, as shown in srfr1-4, a mutation in SRFR1 in Col-0 leads to extreme stunting and abnormal growth because of the constitutive activation of the Col-0-specific resistance gene SNC1 [13]. G1-slsrfr1 plants showed weak growth reduction, not severe stunting, reminiscent of RLD srfr1 phenotype (Figure 2a). As shown in Figure 2b, like Arabidopsis srfr1 mutants [2], SlPR1 and SlPR2 expression were significantly increased in two independent G1-slsrfr1 mutants, slsrfr1-1 and slsrfr1-2, compared to wild-type M82. Consistent with this, SlPR1 protein was strongly accumulated in G1-slsrfr1-1 lines (Figure 2c). These suggest that CRISPR/Cas9-mediated mutations in SlSRFR1 upregulate the expression level of SA-dependent defense markers both transcriptionally and translationally. In addition, consistent with atsrfr1, TomloxD, a JA signaling marker gene, was induced in slsrfr1 mutants (Figure 2b) [7], suggesting both SA- and JA-dependent defense is upregulated in untreated CRISPR/Cas9-edited slsrfr1 lines.
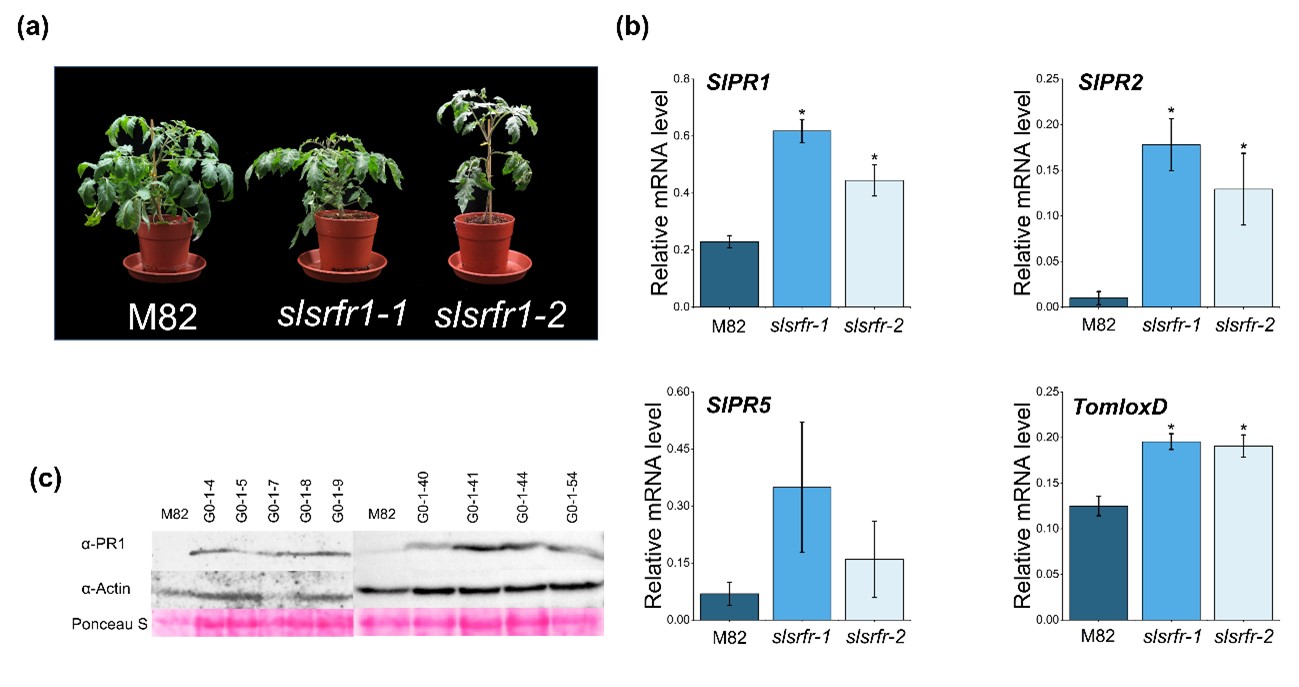
Figure 2. Altered morphology and expression of defense marker genes in CRISPR/Cas9-edited slsrfr1 plants. (a) Growth phenotype of 6-week-old M82 and slsrfr1 lines grown in 16 h light/8 h dark long-day photoperiod; (b) Relative mRNA expression of defense-related genes. The genes used for gene expression analysis refer to SlPR1 (Solyc09g007010.1), SlPR2 (Solyc01g008620.2), SlPR5(Solyc08g080640), and TomloxD (Solyc03g122340.2). Gene expression levels of each gene were normalized with SlACT (Solyc04g011500.3.1) as an internal control. Error bars represent standard deviation. A statistically significant difference was determined by the Student’s t-test (P<0.01). This experiment was repeated twice with similar results; (c) Proteins were extracted from 6-week-old slsrfr1-1 G1 plants and were detected by western blot using α-PR1 and α-Actin (an internal control).
4. Enhanced Resistance to Pto DC3000 in CRISPR/Cas9-Edited slsrfr1 Plants
The absence of increased resistance to virulent Pto DC3000 is observed in Arabidopsis srfr1 mutants, even though defense-related genes are constitutively upregulated [2][13]. Both CRISPR/Cas9-Edited slsrfr1-1 and slsrfr1-2 leaves showed enhanced resistance to Pto DC3000 in contrast to wild-type M82. Disease symptoms in M82, such as chlorosis and water-soaked lesions, were dramatically reduced in G1- slsrfr1 lines (Figure 3a). Consistent with the visible symptoms, slsrfr1-1 and slsrfr1-2 showed approximately 100-fold lower Pto DC3000 growth than wild-type M82 (Figure 3b). These results suggest that mutations in SlSRFR1 increased SA-pathway defense genes, leading to enhanced resistance against the (hemi-)biotrophic pathogen Pto DC3000.
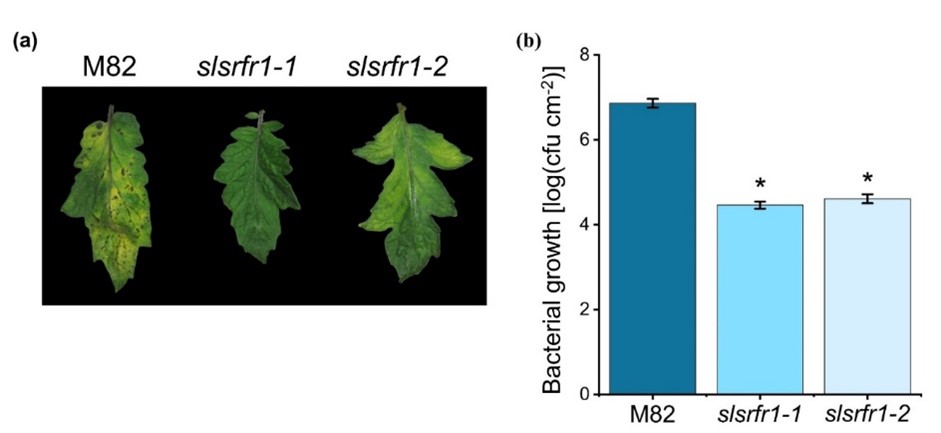
Figure 3. Response of slsrfr1 lines to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000. (a) Disease symptoms of parental M82 (left), slsrfr1-1 (middle), and slsrfr1-2 (right) dip-inoculated with Pto DC3000. Disease symptoms (leaf chlorosis) were recorded at 5 DPI. Only the fourth trifoliate leaflet of the plants was inoculated; (b) In planta bacterial growth was measured in indicated plant lines on day 5 after inoculation with Pto DC3000 at a density of 2 × 108 cfu/mL. Values represent averages of cfu/cm2 leaf tissue from 4 replicas, and error bars denote standard deviation. Asterisks indicate that the growth of DC3000 was significantly different between M82 and slsrfr1 mutants as determined by a two-tailed Student’s t-test (* P < 0.01). This experiment was repeated twice with similar results.
5. Enhanced Susceptibility to Fusarium oxysporum f. sp. lycopersici in CRISPR/Cas9-Edited slsrfr1 Plants
As shown in Figure 1, the Arabidopsis RLD srfr1 mutants display enhanced susceptibility against FOL and B. cinerea. To test the functional conservation of SRFR1 in tomato, we analyzed the resistance response of G1-slsrfr1 lines after inoculation with the necrotrophic fungal pathogen FOL. Three days after plug-inoculation with FOL, all slsrfr1 mutants, slsrfr1-1, slsrfr1-2, slsrfr1-3, and slsrfr1-4, displayed enhanced susceptibility compared to wild-type M82, as indicated by severe necrosis and enlarged lesion area (Figure 4a and 4c). Trypan blue staining of the infected leaves showed that extensive development of fungal hyphae was observed in slsrfr1-1 and slsrfr1-2 with expanding lesion areas (Figure 8b). These results demonstrate that SlSRFR1 positively regulates the immune response against the necrotrophic pathogen FOL, and that SRFR1 function is conserved between Arabidopsis and tomato.
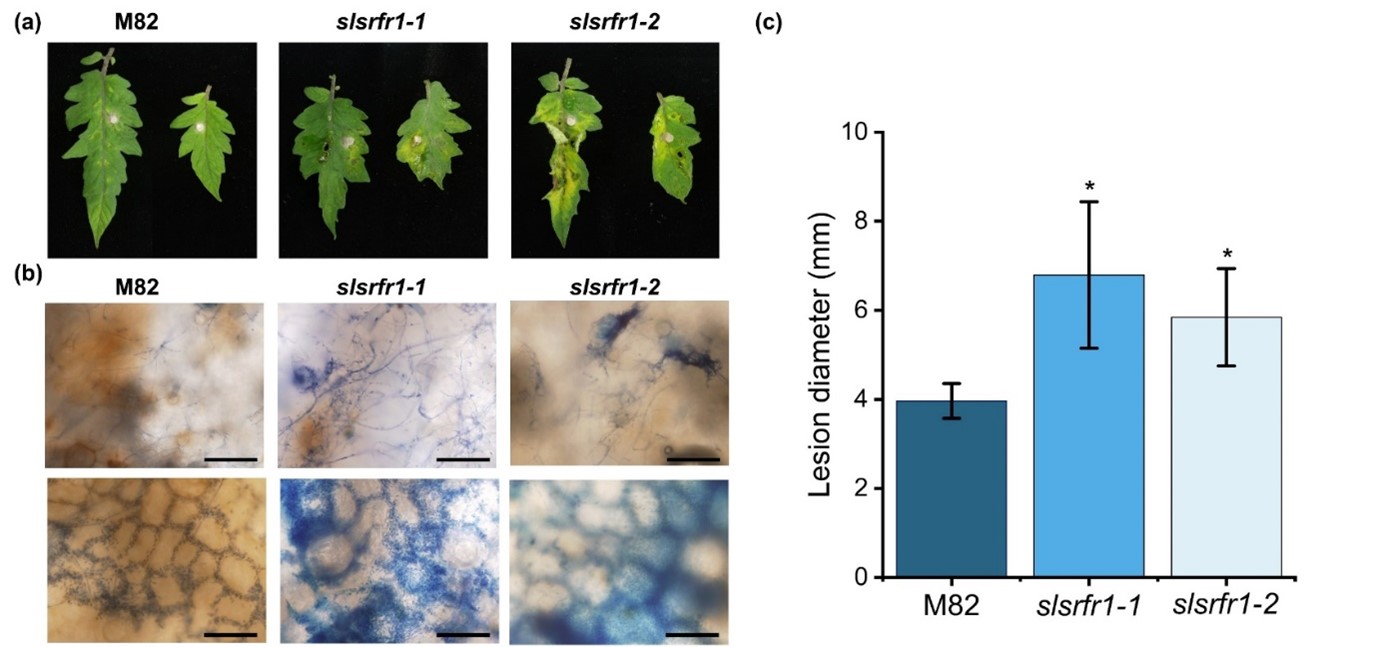
Figure 4. Plant response to Fusarium oxysporum f. sp. lycopersici in slsrfr1 lines. Plant response was analyzed at 3 days after plug inoculation of FOL in slsrfr1 lines. (a) Detached leaves from 6-week-old M82, slsrfr1-1, and slsrfr1-2 were inoculated with 4 mm-diameter plugs of F. oxysproum. Photographs were taken at 3 DPI; (b) Trypan blue staining in FOL-inoculated slsrfr1 lines. The bar represents 200 μm in the upper layer and 100 μm in the lower layer; (c) Lesion size at 3 DPI in slsrfr1 lines. Plant response of slsrfr1-1 and slsrfr1-2 against FOL was repeated three times and once, respectively, with similar results. A statistically significant difference was determined by the Student’s t-test (P<0.01).
6. Current Insights
References
- Kwon, S.I.; Kim, S.H.; Bhattacharjee, S.; Noh, J.J.; Gassmann, W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009, 57, 109–119.
- Kim, S.H.; Kwon, S.I.; Bhattacharjee, S.; Gassmann, W. Regulation of defense gene expression by Arabidopsis SRFR1. Plant Signal. Behav. 2009, 4, 149–150.
- Kim, S.H.; Kwon, S.I.; Saha, D.; Anyanwu, N.C.; Gassmann, W. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 2009, 150, 1723–1732.
- Kwon, S.I.; Koczan, J.M.; Gassmann, W. Two Arabidopsis srfr (suppressor of rps4-RLD) mutants exhibit avrRps4-specific disease resistance independent of RPS4. Plant J. 2004, 40, 366–375.
- Li, Y.; Li, S.; Bi, D.; Cheng, Y.T.; Li, X.; Zhang, Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010, 6, e1001111.
- Kim, S.H.; Son, G.H.; Bhattacharjee, S.; Kim, H.J.; Nam, J.C.; Nguyen, P.D.; Hong, J.C.; Gassmann, W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014, 78, 978–989.
- Nguyen, P.D.; Pike, S.; Wang, J.; Nepal Poudel, A.; Heinz, R.; Schultz, J.C.; Koo, A.J.; Mitchum, M.G.; Appel, H.M.; Gassmann, W. The Arabidopsis immune regulator SRFR1 dampens defences against herbivory by Spodoptera exigua and parasitism by Heterodera schachtii. Mol. Plant Pathol. 2016, 17, 588–600.
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science (New York, N.Y 2011, 334, 1405-1408, doi:10.1126/science.1211592.
- Takken, F.; Rep, M. The arms race between tomato and Fusarium oxysporum. Molecular plant pathology 2010, 11, 309-314, doi:10.1111/j.1364-3703.2009.00605.x.
- Thatcher, L.F.; Gardiner, D.M.; Kazan, K.; Manners, J.M. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol Plant Microbe Interact 2012, 25, 180-190, doi:10.1094/MPMI-08-11-0212.
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; van Kan, J.A. Botrytis cinerea: the cause of grey mould disease. Molecular plant pathology 2007, 8, 561-580, doi:10.1111/j.1364-3703.2007.00417.x.
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. The Plant cell 2012, 24, 3530-3557, doi:10.1105/tpc.112.102046.
- Kim, S.H.; Gao, F.; Bhattacharjee, S.; Adiasor, J.A.; Nam, J.C.; Gassmann, W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS pathogens 2010, 6, e1001172, doi:10.1371/journal.ppat.1001172.
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual review of phytopathology 2005, 43, 205-227, doi:10.1146/annurev.phyto.43.040204.135923.
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proceedings of the National Academy of Sciences of the United States of America 2007, 104, 18842-18847.
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. The Plant cell 2006, 18, 257-273, doi:tpc.105.035576 [pii]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. The Plant cell 2003, 15, 2551-2565, doi:10.1105/tpc.014167.
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant physiology 2012, 159, 266-285, doi:10.1104/pp.111.192641.
- Rahman, T.A.; Oirdi, M.E.; Gonzalez-Lamothe, R.; Bouarab, K. Necrotrophic pathogens use the salicylic acid signaling pathway to promote disease development in tomato. Mol Plant Microbe Interact 2012, 25, 1584-1593, doi:10.1094/MPMI-07-12-0187-R.
- Hernandez-Aparicio, F.; Lison, P.; Rodrigo, I.; Belles, J.M.; Lopez-Gresa, M.P. Signaling in the Tomato Immunity against Fusarium oxysporum. Molecules 2021, 26, doi:10.3390/molecules26071818.
- Garner, C.M.; Spears, B.J.; Su, J.; Cseke, L.J.; Smith, S.N.; Rogan, C.J.; Gassmann, W. Opposing functions of the plant TOPLESS gene family during SNC1-mediated autoimmunity. PLoS genetics 2021, 17, e1009026, doi:10.1371/journal.pgen.1009026.




