
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Piotr Bednarek | + 4465 word(s) | 4465 | 2021-08-04 08:55:46 | | | |
| 2 | Lily Guo | + 27 word(s) | 4492 | 2021-08-06 03:15:26 | | |
Video Upload Options
Somaclonal variation includes genetic or epigenetic changes exhibited between clonal regenerants and their corresponding donor plants derived via in vitro tissue cultures (A. Leva, L.M.R. Rinaldi, in Encyclopedia of Applied Plant Sciences (Second Edition), 2017). It usually assumes that the changes are being transmitted during a generative cycle. However, in some cases, to stress the fact that not all changes are either not analyzed in the progeny or may not be sexually transmitted, the tissue culture-induced variation seems to reflect better the issue (Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.) Bednarek, PT., Orłowska, R., Koebner, RMD., Zimny, J. 2007 BMC Plant Biology 7 (1), 1-9).
1. Introduction
2. Stresses and Their Role in Plant Regeneration through Tissue Culture In Vitro
3. Cell Wall and Plasma Membrane as Sensors of Stresses
4. Retrograde and Anterograde Signaling
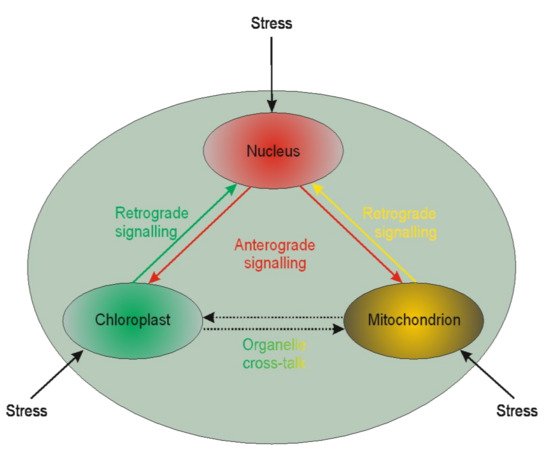
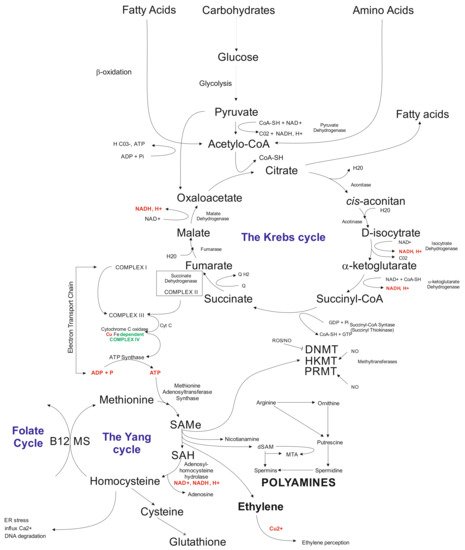
5. Biochemical Aspects
References
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol. Biol. 2020, 103, 33–50.
- Machczyńska, J.; Zimny, J.; Bednarek, P. Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol. Biol. 2015, 89, 279–292.
- Mikuła, A.; Tomiczak, K.; Rybczynski, J.J. Cryopreservation enhances embryogenic capacity of Gentiana cruciata (L.) suspension culture and maintains (epi)genetic uniformity of regenerants. Plant Cell Rep. 2011, 30, 565–574.
- Fiuk, A.; Bednarek, P.T.; Rybczyński, J.J. Flow Cytometry, HPLC-RP, and metAFLP Analyses to Assess Genetic Variability in Somatic Embryo-Derived Plantlets of Gentiana pannonica Scop. Plant Mol. Biol. Rep. 2010, 28, 413–420.
- Agarwal, P.K.; Bhojwani, S.S. Genetic variability in the progeny of androgenic dihaploid plants and selection of high agronomic performing lines in Brassica juncea. Biol. Plant. 2004, 48, 503–508.
- Zehr, B.E.; Williams, M.E.; Duncan, D.R.; Widholm, J.M. Somaclonal variation in the progeny of plants regenerated from callus cultures of seven inbred lines of maize. Can. J. Bot. 1987, 65, 491–499.
- Ghosh, A.; Igamberdiev, A.U.; Debnath, S.C. Tissue culture-induced DNA methylation in crop plants: A review. Mol. Biol. Rep. 2021, 48, 823–841.
- Cao, Z.; Sui, S.; Cai, X.; Yang, Q.; Deng, Z. Somaclonal variation in ‘Red Flash’ caladium: Morphological, cytological and molecular characterization. Plant Cell Tissue Organ Cult. 2016, 126, 269–279.
- Deepthi, V.P. Somaclonal variation in micro propagated bananas. Adv. Plants Agric. Res. 2018, 8, 624–627.
- Qin, Y.; Shin, K.-S.; Woo, H.-J.; Lim, M.-H. Genomic Variations of Rice Regenerants from Tissue Culture Revealed by Whole Genome Re-Sequencing. Plant Breed. Biotechnol. 2018, 6, 426–433.
- Azizi, P.; Hanafi, M.M.; Sahebi, M.; Harikrishna, J.A.; Taheri, S.; Yassoralipour, A.; Nasehi, A. Epigenetic changes and their relationship to somaclonal variation: A need to monitor the micropropagation of plantation crops. Funct. Plant Biol. 2020, 47, 508–523.
- Orłowska, R. Barley somatic embryogenesis-an attempt to modify variation induced in tissue culture. J. Biol. Res. Thessalon. 2021, 28, 9.
- Skirvin, R.M.; Janick, J. Tissue culture-induced variation in scented Pelargonium spp. J. Am. Soc. Hortic. Sci. 1976, 101, 281–290.
- Scowcroft, W.R. Somaclonal variation: The myth of clonal uniformity. In Genetic Flux in Plants; Hohn, B., Dennis, E.B., Eds.; Springer: Vienna, Austria, 1985; pp. 217–245.
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvment. Theor. Appl. Genet. 1981, 60, 197–214.
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331.
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polimorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535.
- Jeong, I.S.; Yoon, U.H.; Lee, G.S.; Ji, H.S.; Lee, H.J.; Han, C.D.; Hahn, J.H.; An, G.; Kim, T.H. SNP-based analysis of genetic diversity in anther-derived rice by whole genome sequencing. Rice 2013, 6, 6.
- Peschke, V.M.; Phillips, R.L.; Genggenbach, B.G. Discovery of transposable element activity among progeny of tissue-culture derived plants. Science 1987, 238, 804–807.
- Sato, M.; Hosokawa, M.; Doi, M. Somaclonal Variation Is Induced De Novo via the Tissue Culture Process: A Study Quantifying Mutated Cells in Saintpaulia. PLoS ONE 2011, 6, e23541.
- Barret, P.; Brinkman, M.; Beckert, M. A sequence related to rice Pong transposable element displays transcriptional activation by in vitro culture and reveals somaclonal variations in maize. Genome 2006, 49, 1399–1407.
- Tanurdzic, M.; Vaughn, M.W.; Jiang, H.; Lee, T.-J.; Slotkin, R.K.; Sosinski, B.; Thompson, W.F.; Doerge, R.W.; Martienssen, R.A. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008, 6, e302.
- Kaeppler, S.M.; Phillips, R.L.; Olhoft, P. Molecular basis of heritable tissue culture-induced variation in plants. In Somaclonal Variation and Induced Mutation in Crop Improvement; Jain, S.M., Brar, D.S., Ahloowalia, B.S., Eds.; Kluwer Academic: Dordrecht, The Netherlands, 1998; pp. 465–484.
- Cassels, A.C.; Curry, R.F. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: Implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult. 2001, 64, 145–157.
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54.
- Dey, T.; Saha, S.; Ghosh, P.D. Somaclonal variation among somatic embryo derived plants—Evaluation of agronomically important somaclones and detection of genetic changes by RAPD in Cymbopogon winterianus. S. Afr. J. Bot. 2015, 96, 112–121.
- Touraev, A.; Indrianto, A.; Wratschko, I.; Vicente, O.; Heberle-Bors, E. Efficient microspore embryogenesis in wheat (Triticum aestivum L.) induced by starvation at high temperature. Sex. Plant Reprod. 1996, 9, 209–215.
- Hoekstra, S.; Hoekstra, S.; Hoekstra, I.R.; Hoekstra, R.A.; Hoekstra, E. The Interaction of 2,4-D Application and Mannitol Pretreatment in Anther and Microspore Culture of Hordeum vulgare L. cv. Igri. J. Plant Physiol. 1996, 148, 696–700.
- Binarova, P.; Hause, G.; Cenklová, V.; Cordewener, J.H.; Campagne, M.L. A short severe heat shock is required to induce embryogenesis in late bicellular pollen of Brassica napus L. Sex. Plant Reprod. 1997, 10, 200–208.
- Popova, T.; Grozeva, S.; Todorova, V.; Stankova, G.; Anachkov, N.; Rodeva, V. Effects of low temperature, genotype and culture media on in vitro androgenic answer of pepper (Capsicum annuum L.). Acta Physiol. Plant. 2016, 38, 273.
- Tenhola-Roininen, T.; Tanhuanpää, P.; Immonen, S. The effect of cold and heat treatments on the anther culture response of diverse rye genotypes. Euphytica 2005, 145, 1–9.
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414.
- Reyna-Lopez, G.E.; Simpson, J.; Ruiz-Herrera, J. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 1997, 253, 703–710.
- Xiong, L.Z.; Xu, C.G.; Saghai Maroof, M.A.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplifcation polymorphism technique. Mol. Genet. Genom. 1999, 261, 439–446.
- Baranek, M.; Cechova, J.; Kovacs, T.; Eichmeier, A.; Wang, S.; Raddova, J.; Necas, T.; Ye, X. Use of Combined MSAP and NGS Techniques to Identify Differentially Methylated Regions in Somaclones: A Case Study of Two Stable Somatic Wheat Mutants. PLoS ONE 2016, 11, e0165749.
- Francischini, J.H.M.B.; Kemper, E.L.; Costa, J.B.; Manechini, J.R.V.; Pinto, L.R. DNA methylation in sugarcane somaclonal variants assessed through methylation-sensitive amplified polymorphism. Genet. Mol. Res. 2017, 16.
- Bobadilla Landey, R.; Cenci, A.; Georget, F.; Bertrand, B.; Camayo, G.; Dechamp, E.; Herrera, J.C.; Santoni, S.; Lashermes, P.; Simpson, J.; et al. High Genetic and Epigenetic Stability in Coffea arabica Plants Derived from Embryogenic Suspensions and Secondary Embryogenesis as Revealed by AFLP, MSAP and the Phenotypic Variation Rate. PLoS ONE 2013, 8, e56372.
- Dann, A.L.; Wilson, C.R. Comparative assessment of genetic and epigenetic variation among regenerants of potato (Solanum tuberosum) derived from long-term nodal tissue-culture and cell selection. Plant Cell Rep. 2011, 30, 631–639.
- Guevara, M.; de María, N.; Sáez-Laguna, E.; Vélez, M.D.; Cervera, M.T.; Cabezas, J.A. Analysis of DNA Cytosine Methylation Patterns Using Methylation-Sensitive Amplification Polymorphism (MSAP). Methods Mol. Biol. 2017, 1456, 99–112.
- Fulneček, J.; Kovařík, A. How to interpret Methylation Sensitive Amplified Polymorphism (MSAP) profiles? BMC Genet. 2014, 15, 2.
- Alonso, C.; Balao, F.; Bazaga, P.; Perez, R. Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. 2016, 212, 571–576.
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 13, 642–653.
- Bednarek, P.T.; Orłowska, R.; Niedziela, A. A relative quantitative Methylation-Sensitive Amplified Polymorphism (MSAP) method for the analysis of abiotic stress. BMC Plant Biol. 2017, 17, 79.
- Machczyńska, J.; Orłowska, R.; Zimny, J.; Bednarek, P.T. Extended metAFLP approach in studies of the tissue culture induced variation (TCIV) in case of triticale. Mol. Breed. 2014, 34, 845–854.
- Bednarek, P.T.; Orłowska, R. Time of In Vitro Anther Culture May Moderate Action of Copper and Silver Ions that Affect the Relationship between DNA Methylation Change and the Yield of Barley Green Regenerants. Plants 2020, 9, 1064.
- Wang, S.; Lv, J.; Zhang, L.; Dou, J.; Sun, Y.; Li, X.; Fu, X.; Dou, H.; Mao, J.; Hu, X.; et al. MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 2015, 5, 150130.
- Brunner, A.L.; Johnson, D.S.; Kim, S.W.; Valouev, A.; Reddy, T.E.; Neff, N.F.; Anton, E.; Medina, C.; Nguyen, L.; Chiao, E.; et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009, 19, 1044–1056.
- Haque, N.; Nishiguchi, M. Bisulfite sequencing for cytosine-methylation analysis in plants. Methods Mol. Biol. 2011, 744, 187–197.
- Zhang, Y.; Rohde, C.; Tierling, S.; Stamerjohanns, H.; Reinhardt, R.; Walter, J.; Jeltsch, A. DNA Methylation Analysis by Bisulfite Conversion, Cloning, and Sequencing of Individual Clones. In DNA Methylation: Methods and Protocols; Tost, J., Ed.; Humana Press: Totowa, NY, USA, 2009; pp. 177–187.
- Sun, Q.; Qiao, J.; Zhang, S.; He, S.; Shi, Y.; Yuan, Y.; Zhang, X.; Cai, Y. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development. PeerJ 2018, 6, e4945.
- Bednarek, P.T.; Orłowska, R.; Koebner, R.M.D.; Zimny, J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol. 2007, 7, 10.
- Orłowska, R.; Machczyńska, J.; Oleszczuk, S.; Zimny, J.; Bednarek, P.T. DNA methylation changes and TE activity induced in tissue cultures of barley (Hordeum vulgare L.). J. Biol. Res. 2016, 23, 19.
- Machczyńska, J.; Orłowska, R.; Mańkowski, D.R.; Zimny, J.; Bednarek, P.T. DNA methylation changes in triticale due to in vitro culture plant regeneration and consecutive reproduction. Plant Cell Tissue Organ Cult. 2014, 119, 289–299.
- Śliwińska, A.A.; Białek, A.; Orłowska, R.; Mańkowski, D.; Sykłowska-Baranek, K.; Pietrosiuk, A. Comparative Study of the Genetic and Biochemical Variability of Polyscias filicifolia (Araliaceae) Regenerants Obtained by Indirect and Direct Somatic Embryogenesis as a Source of Triterpenes. Int. J. Mol. Sci. 2021, 22, 5752.
- Mikuła, A.; Tomiczak, K.; Wójcik, A.; Rybczynski, J.J. Encapsulation-dehydration method elevates embryogenic abilities of Gentiana kurroo cell suspension and carrying on genetic stability of its regenerants after cryopreservation. In Acta Horticulturae; International Society for Horticultural Science: Leuven, Belgium, 2011; Volume 908, pp. 143–154.
- Oleszczuk, S.; Zimny, J.; Bednarek, P.T. The application of the AFLP method to determine the purity of homozygous lines of barley (Hordeum vulgare L.). Cell. Mol. Biol. Lett. 2002, 7, 777–783.
- Coronel, C.J.; González, A.I.; Ruiz, M.L.; Polanco, C. Analysis of somaclonal variation in transgenic and regenerated plants of Arabidopsis thaliana using methylation related metAFLP and TMD markers. Plant Cell Rep. 2018, 37, 137–152.
- Lukens, L.N.; Zhan, S. The plant genome’s methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 2007, 10, 317–322.
- Jiang, C.; Mithani, A.; Gan, X.; Belfield, E.J.; Klingler, J.P.; Zhu, J.K.; Ragoussis, J.; Mott, R.; Harberd, N.P. Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr. Biol. 2011, 21, 1385–1390.
- Orłowska, R.; Pachota, K.A.; Dynkowska, W.M.; Niedziela, A.; Bednarek, P.T. Androgenic-Induced Transposable Elements Dependent Sequence Variation in Barley. Int. J. Mol. Sci. 2021, 22, 6783.
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791.
- Kumar, S.; Lahlali, R.; Liu, X.; Karunakaran, C. Infrared spectroscopy combined with imaging: A new developing analytical tool in health and plant science. Appl. Spectrosc. Rev. 2016, 51, 466–483.
- Kačuráková, M.; Capek, P.; Sasinková, V.; Wellner, N.; Ebringerová, A. FT-IR study of plant cell wall model compounds: Pectic polysaccharides and hemicelluloses. Carbohydr. Polym. 2000, 43, 195–203.
- Kazarian, S.G.; Chan, K.L.A. ATR-FTIR spectroscopic imaging: Recent advances and applications to biological systems. Analyst 2013, 138, 1940–1951.
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann. Transl. Med. 2014, 2, 17.
- Lou, Y.; Zhu, J.; Yang, Z. Molecular Cell Biology of Pollen Walls. In Applied Plant Cell Biology: Cellular Tools and Approaches for Plant Biotechnology; Nick, P., Opatrny, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 179–205.
- Bednarek, P.T.; Zebrowski, J.; Orłowska, R. Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture. Int. J. Mol. Sci. 2020, 21, 5770.
- Bednarek, P.T.; Orłowska, R. CG Demethylation Leads to Sequence Mutations in an Anther Culture of Barley Due to the Presence of Cu, Ag Ions in the Medium and Culture Time. Int. J. Mol. Sci. 2020, 21, 4401.
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis. A Regression Bases Approach; A Division of Guilford Publications, Inc.: New York, NY, USA, 2018; p. 507.
- Maraschin, S.F.; de Priester, W.; Spaink, H.P.; Wang, M. Androgenic switch: An example of plant embryogenesis from the male gametophyte perspective. J. Exp. Bot. 2005, 56, 1711–1726.
- Galán-Ávila, A.; García-Fortea, E.; Prohens, J.; Herraiz, F.J. Microgametophyte Development in Cannabis sativa L. and First Androgenesis Induction Through Microspore Embryogenesis. Front. Plant Sci. 2021, 12, 669424.
- Testillano, P.S. Microspore embryogenesis: Targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 2019, 70, 2965–2978.
- Seguí-Simarro, J.M.; Bárány, I.; Suárez, R.; Fadón, B.; Testillano, P.S.; Risueño, M.C. Nuclear bodies domain changes with microspore reprogramming to embryogenesis. Eur. J. Histochem. 2006, 50, 35–44.
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176.
- Rodríguez-Serrano, M.; Bárány, I.; Prem, D.; Coronado, M.-J.; Risueño, M.C.; Testillano, P.S. NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 2012, 63, 2007–2024.
- Pasternak, T. Oxidative stress inducing agents’ copper and alloxan accelerate cell cycle re-entering of somatic plant cells in the presence of suboptimal exogenous auxin. bioRxiv 2020. preprint.
- Uváčková, L.; Takáč, T.; Boehm, N.; Obert, B.; Samaj, J. Proteomic and biochemical analysis of maize anthers after cold pretreatment and induction of androgenesis reveals an important role of anti-oxidative enzymes. J. Proteom. 2012, 75, 1886–1894.
- Da Costa, C.; De Almeida, M.; Ruedell, C.; Schwambach, J.; Maraschin, F.; Fett-Neto, A. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133.
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta 2015, 1849, 385–402.
- Pasternak, T.P.; Prinsen, E.; Ayaydin, F.; Miskolczi, P.l.; Potters, G.; Asard, H.; Van Onckelen, H.A.; Dudits, D.n.; Fehér, A. The Role of Auxin, pH, and Stress in the Activation of Embryogenic Cell Division in Leaf Protoplast-Derived Cells of Alfalfa. Plant Physiol. 2002, 129, 1807–1819.
- Pasternak, T.P.; Ötvös, K.; Domoki, M.; Fehér, A. Linked activation of cell division and oxidative stress defense in alfalfa leaf protoplast-derived cells is dependent on exogenous auxin. Plant Growth Regul. 2007, 51, 109–117.
- Rodríguez-Sanz, H.; Solís, M.T.; López, M.F.; Gómez-Cadenas, A.; Risueño, M.C.; Testillano, P.S. Auxin Biosynthesis, Accumulation, Action and Transport are Involved in Stress-Induced Microspore Embryogenesis Initiation and Progression in Brassica napus. Plant Cell Physiol. 2015, 56, 1401–1417.
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136.
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47.
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The Systems Biology of Auxin in Developing Embryos. Trends Plant Sci. 2017, 22, 225–235.
- Su, Y.H.; Liu, Y.B.; Bai, B.; Zhang, X.S. Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front. Plant Sci. 2015, 5, 792.
- Hofius, D.; Li, L.; Hafrén, A.; Coll, N.S. Autophagy as an emerging arena for plant-pathogen interactions. Curr. Opin. Plant Biol. 2017, 38, 117–123.
- Masclaux-Daubresse, C.; Chen, Q.; Havé, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 2017, 39, 8–17.
- Avin-Wittenberg, T.; Baluška, F.; Bozhkov, P.V.; Elander, P.H.; Fernie, A.R.; Galili, G.; Hassan, A.; Hofius, D.; Isono, E.; Le Bars, R.; et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Experimrntal Bot. 2018, 69, 1335–1353.
- Corral-Martínez, P.; Parra-Vega, V.; Seguí-Simarro, J.M. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: Evidence for massive autophagy and excretion-based cytoplasmic cleaning. J. Exp. Bot. 2013, 64, 3061–3075.
- Michaeli, S.; Avin-Wittenberg, T.; Galili, G. Involvement of autophagy in the direct ER to vacuole protein trafficking route in plants. Front. Plant Sci. 2014, 5, 134.
- Tan, X.; Li, K.; Wang, Z.; Zhu, K.; Tan, X.; Cao, J. A Review of Plant Vacuoles: Formation, Located Proteins, and Functions. Plants 2019, 8, 327.
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Reactive Oxygen Species and Autophagy in Plants and Algae. Plant Physiol. 2012, 160, 156–164.
- Bárány, I.; Berenguer, E.; Solís, M.T.; Pérez-Pérez, Y.; Santamaría, M.E.; Crespo, J.L.; Risueño, M.C.; Díaz, I.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402.
- Tsiatsiani, L.; Van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288.
- Berenguer, E.; Bárány, I.; Solís, M.-T.; Pérez-Pérez, Y.; Risueño, M.C.; Testillano, P.S. Inhibition of Histone H3K9 Methylation by BIX-01294 Promotes Stress-Induced Microspore Totipotency and Enhances Embryogenesis Initiation. Front. Plant Sci. 2017, 8, 161.
- Solís, M.-T.; El-Tantawy, A.-A.; Cano, V.; Risueño, M.C.; Testillano, P.S. 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front. Plant Sci. 2015, 6, 472.
- El-Tantawy, A.A.; Solís, M.T.; Risueño, M.C.; Testillano, P.S. Changes in DNA Methylation Levels and Nuclear Distribution Patterns after Microspore Reprogramming to Embryogenesis in Barley. Cytogenet. Genome Res. 2014, 143, 200–208.
- Çakmak, E.; Uncuoğlu, A.A.; Aydın, Y. Evaluation of in vitro genotoxic effects induced by in vitro anther culture conditions in sunflower. Plant Signal. Behav. 2019, 14, 1633885.
- Zieliński, K.; Krzewska, M.; Żur, I.; Juzoń, K.; Kopeć, P.; Nowicka, A.; Moravčiková, J.; Skrzypek, E.; Dubas, E. The effect of glutathione and mannitol on androgenesis in anther and isolated microspore cultures of rye (Secale cereale L.). Plant Cell Tissue Organ Cult. 2020, 140, 577–592.
- Castander-Olarieta, A.; Montalbán, I.A.; De Medeiros Oliveira, E.; Dell’Aversana, E.; D’Amelia, L.; Carillo, P.; Steiner, N.; Fraga, H.P.D.F.; Guerra, M.P.; Goicoa, T.; et al. Effect of Thermal Stress on Tissue Ultrastructure and Metabolite Profiles During Initiation of Radiata Pine Somatic Embryogenesis. Front. Plant Sci. 2019, 9, 4.
- Gamalero, E.; Glick, B.R. Ethylene and Abiotic Stress Tolerance in Plants. In Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Ahmad, P., Prasad, M.N.V., Eds.; Springer New York: New York, NY, USA, 2012; pp. 395–412.
- Klay, I.; Pirrello, J.; Riahi, L.; Bernadac, A.; Cherif, A.; Bouzayen, M.; Bouzid, S. Ethylene response factor Sl-ERF. B. 3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci. World J. 2014, 2014, 167681.
- Mu, C.; Wang, S.; Zhang, S.; Pan, J.; Chen, N.; Li, X.; Wang, Z.; Liu, H. Small heat shock protein LimHSP16.45 protects pollen mother cells and tapetal cells against extreme temperatures during late zygotene to pachytene stages of meiotic prophase I in David Lily. Plant Cell Rep. 2011, 30, 1981.
- Orłowska, R.; Pachota, K.A.; Machczyńska, J.; Niedziela, A.; Makowska, K.; Zimny, J.; Bednarek, P.T. Improvement of anther cultures conditions using the Taguchi method in three cereal crops. Electron. J. Biotechnol. 2020, 43, 8–15.
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932.
- De Lorenzo, G.; Ferrari, S.; Giovannoni, M.; Mattei, B.; Cervone, F. Cell wall traits that influence plant development, immunity, and bioconversion. Plant J. 2019, 97, 134–147.
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460.
- Quilichini, T.D.; Grienenberger, E.; Douglas, C.J. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 2015, 113, 170–182.
- Heslop-Harrison, J. Pollen wall development. Science 1968, 161, 230–237.
- Blackmore, S.; Wortley, A.H.; Skvarla, J.J.; Rowley, J.R. Pollen wall development in flowering plants. New Phytol. 2007, 174, 483–498.
- Ahlers, F.; Lambert, J.; Wiermann, R. Acetylation and silylation of piperidine solubilized sporopollenin from pollen of Typha angustifolia L. Z. Für Nat. C 2003, 58, 807–811.
- Blokker, P.; Boelen, P.; Broekman, R.; Rozema, J. The occurrence of p-coumaric acid and ferulic acid in fossil plant materials and their use as UV-proxy. In Plants and Climate Change; Springer: Berlin/Heidelberg, Germany, 2006; pp. 197–208.
- Bubert, H.; Lambert, J.; Steuernagel, S.; Ahlers, F.; Wiermann, R. Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Verl. Der Z. Für Nat. 2002, 57, 1035–1041.
- Li, W.L.; Liu, Y.; Douglas, C.J. Role of Glycosyltransferases in Pollen Wall Primexine Formation and Exine Patterning. Plant Physiol. 2017, 173, 167–182.
- Heslop-Harrison, J. Wall development within the microspore tetrad of Lilium longiflorum. Can. J. Bot. 1968, 46, 1185–1192.
- Quilichini, T.D.; Douglas, C.J.; Samuels, A.L. New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Ann. Bot. 2014, 114, 1189–1201.
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771.
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113.
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131.
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951.
- Saffer, A.M. Expanding roles for pectins in plant development. J. Integr. Plant Biol. 2018, 60, 910–923.
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296.
- Barnes, W.J.; Anderson, C.T. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J. 2018, 94, 956–974.
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94.
- Kurek, I.; Kawagoe, Y.; Jacob-Wilk, D.; Doblin, M.; Delmer, D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc. Natl. Acad. Sci. USA 2002, 99, 11109–11114.
- Sethaphong, L.; Haigler, C.H.; Kubicki, J.D.; Zimmer, J.; Bonetta, D.; DeBolt, S.; Yingling, Y.G. Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. USA 2013, 110, 7512–7517.
- Slabaugh, E.; Davis, J.K.; Haigler, C.H.; Yingling, Y.G.; Zimmer, J. Cellulose synthases: New insights from crystallography and modeling. Trends Plant Sci. 2014, 19, 99–106.
- Hill, J.L.; Hammudi, M.B.; Tien, M. The Arabidopsis cellulose synthase complex: A proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 2014, 26, 4834–4842.
- Napoleão, T.A.; Soares, G.; Vital, C.E.; Bastos, C.; Castro, R.; Loureiro, M.E.; Giordano, A. Methyl jasmonate and salicylic acid are able to modify cell wall but only salicylic acid alters biomass digestibility in the model grass Brachypodium distachyon. Plant Sci. 2017, 263, 46–54.
- Wang, Y.; Leng, L.; Islam, M.K.; Liu, F.; Lin, C.S.K.; Leu, S.-Y. Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization. Int. J. Mol. Sci. 2019, 20, 3354.
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832.
- Humphrey, T.V.; Bonetta, D.T.; Goring, D.R. Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 2007, 176, 7–21.
- Kafkaletou, M.; Fasseas, C.; Tsantili, E. Increased firmness and modified cell wall composition by ethylene were reversed by the ethylene inhibitor 1-methylcyclopropene (1-MCP) in the non-climacteric olives harvested at dark green stage—Possible implementation of ethylene for olive quality. J. Plant Physiol. 2019, 238, 63–71.
- Salazar, R.; Pollmann, S.; Morales-Quintana, L.; Herrera, R.; Caparrós-Ruiz, D.; Ramos, P. In seedlings of Pinus radiata, jasmonic acid and auxin are differentially distributed on opposite sides of tilted stems affecting lignin monomer biosynthesis and composition. Plant Physiol. Biochem. 2019, 135, 215–223.
- Hu, Z.; Vanderhaeghen, R.; Cools, T.; Wang, Y.; De Clercq, I.; Leroux, O.; Nguyen, L.; Belt, K.; Millar, A.H.; Audenaert, D.; et al. Mitochondrial Defects Confer Tolerance against Cellulose Deficiency. Plant Cell 2016, 28, 2276–2290.
- Hofmann, N.R. A Functional Link between Mitochondria and the Cell Wall in Stress Responses. Plant Cell 2016, 28, 1996.
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 Coordinates Organellar Functions and Stress Responses by Reprogramming Retrograde Signaling. Plant Physiol. 2019, 180, 634–653.
- Amos, R.A.; Mohnen, D. Critical Review of Plant Cell Wall Matrix Polysaccharide Glycosyltransferase Activities Verified by Heterologous Protein Expression. Front. Plant Sci. 2019, 10, 15.
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575.
- Uluisik, S.; Seymour, G.B. Pectate lyases: Their role in plants and importance in fruit ripening. Food Chem. 2020, 309, 125559.
- Ackermann, F.; Stanislas, T. The Plasma Membrane—An Integrating Compartment for Mechano-Signaling. Plants 2020, 9, 505.
- Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-D-Glucans in Cell Walls of the Poaceae, Lower Plants, and Fungi: A Tale of Two Linkages. Mol. Plant 2009, 2, 873–882.
- Marković, S.M.; Đukić, N.H.; Knežević, D.; Leković, S.V. Divergence of barley and oat varieties according to their content of β-glucan. J. Serb. Chem. Soc. 2017, 82, 379–388.
- Zhai, Z.; Liu, H.; Xu, C.; Shanklin, J. Sugar Potentiation of Fatty Acid and Triacylglycerol Accumulation. Plant Physiol. 2017, 175, 696–707.
- Funck, D.; Winter, G.; Baumgarten, L.; Forlani, G. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 2012, 12, 191.
- Price, J.; Laxmi, A.; St Martin, S.K.; Jang, J.-C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 2004, 16, 2128–2150.
- Jang, J.C.; Sheen, J. Sugar sensing in higher plants. Plant Cell 1994, 6, 1665–1679.
- Pego, J.V.; Smeekens, S.C. Plant fructokinases: A sweet family get-together. Trends Plant Sci. 2000, 5, 531–536.
- Díaz-Sala, C. Molecular Dissection of the Regenerative Capacity of Forest Tree Species: Special Focus on Conifers. Front. Plant Sci. 2019, 9, 1943.
- Gravino, M.; Savatin, D.V.; Macone, A.; De Lorenzo, G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015, 84, 1073–1086.
- Wang, J.; Tergel, T.; Chen, J.; Yang, J.; Kang, Y.; Qi, Z. Arabidopsis transcriptional response to extracellular Ca2+ depletion involves a transient rise in cytosolic Ca2+. J. Integr. Plant Biol. 2015, 57, 138–150.
- Knight, H. Calcium Signaling during Abiotic Stress in Plants. In International Review of Cytology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 1999; Volume 195, pp. 269–324.
- Makowska, K.; Kałużniak, M.; Oleszczuk, S.; Zimny, J.; Czaplicki, A.; Konieczny, R. Arabinogalactan proteins improve plant regeneration in barley (Hordeum vulgare L.) anther culture. Plant Cell Tissue Organ Cult. 2017, 131, 247–257.
- Corral-Martínez, P.; Driouich, A.; Seguí-Simarro, J.M. Dynamic Changes in Arabinogalactan-Protein, Pectin, Xyloglucan and Xylan Composition of the Cell Wall During Microspore Embryogenesis in Brassica napus. Front. Plant Sci. 2019, 10, 332.
- El-Tantawy, A.-A.; Solís, M.-T.; Da Costa, M.L.; Coimbra, S.; Risueño, M.-C.; Testillano, P.S. Arabinogalactan protein profiles and distribution patterns during microspore embryogenesis and pollen development in Brassica napus. Plant Reprod. 2013, 26, 231–243.
- Hernández-Verdeja, T.; Strand, Å. Retrograde Signals Navigate the Path to Chloroplast Development. Plant Physiol. 2017, 176, 967–976.
- Singh, R.; Singh, S.; Parihar, P.; Singh, V.P.; Prasad, S.M. Retrograde signaling between plastid and nucleus: A review. J. Plant Physiol. 2015, 181, 55–66.
- Pfalz, J.; Oelmüller, R. Plastid Retrograde Signals: More to Discover. In Sensory Biology of Plants; Sopory, S., Ed.; Springer: Singapore, 2019; pp. 477–507.
- Fujii, S.; Toriyama, K. Genome Barriers between Nuclei and Mitochondria Exemplified by Cytoplasmic Male Sterility. Plant Cell Physiol. 2008, 49, 1484–1494.
- Börner, T. The discovery of plastid-to-nucleus retrograde signaling—a personal perspective. Protoplasma 2017, 254, 1845–1855.
- Enami, K.; Ozawa, T.; Motohashi, N.; Nakamura, M.; Tanaka, K.; Hanaoka, M. Plastid-to-Nucleus Retrograde Signals Are Essential for the Expression of Nuclear Starch Biosynthesis Genes during Amyloplast Differentiation in Tobacco BY-2 Cultured Cells. Plant Physiol. 2011, 157, 518–530.
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic stress-induced chloroplast proteome remodelling: A mechanistic overview. J. Exp. Bot. 2018, 69, 2773–2781.
- Bélanger, S.; Marchand, S.; Jacques, P.-É.; Meyers, B.; Belzile, F. Differential Expression Profiling of Microspores During the Early Stages of Isolated Microspore Culture Using the Responsive Barley Cultivar Gobernadora. G3 2018, 8, 1603–1614.
- Gajecka, M.; Marzec, M.; Chmielewska, B.; Jelonek, J.; Zbieszczyk, J.; Szarejko, I. Plastid differentiation during microgametogenesis determines green plant regeneration in barley microspore culture. Plant Sci. 2020, 291, 110321.
- Kawanabe, T.; Ariizumi, T.; Kawai-Yamada, M.; Uchimiya, H.; Toriyama, K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 2006, 47, 784–787.
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818.
- Rivas-Sendra, A.; Corral-Martínez, P.; Porcel, R.; Camacho-Fernández, C.; Calabuig-Serna, A.; Seguí-Simarro, J.M. Embryogenic competence of microspores is associated with their ability to form a callosic, osmoprotective subintinal layer. J. Exp. Bot. 2019, 70, 1267–1281.
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154.
- Moffatt, B.A.; Weretilnyk, E.A. Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells. Physiol. Plant. 2001, 113, 435–442.
- Yoshida, T.; Furihata, H.Y.; To, T.K.; Kakutani, T.; Kawabe, A. Genome defense against integrated organellar DNA fragments from plastids into plant nuclear genomes through DNA methylation. Sci. Rep. 2019, 9, 2060.
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533.
- Zierer, W.; Hajirezaei, M.R.; Eggert, K.; Sauer, N.; von Wirén, N.; Pommerrenig, B. Phloem-Specific Methionine Recycling Fuels Polyamine Biosynthesis in a Sulfur-Dependent Manner and Promotes Flower and Seed Development. Plant Physiol. 2016, 170, 790–806.
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945.
- Shang, B.; Xu, C.; Zhang, X.; Cao, H.; Xin, W.; Hu, Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5101–5106.
- Contento, A.L.; Kim, S.-J.; Bassham, D.C. Transcriptome Profiling of the Response of Arabidopsis Suspension Culture Cells to Suc Starvation. Plant Physiol. 2004, 135, 2330–2347.




