
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Saba Shamim | + 4269 word(s) | 4269 | 2021-08-04 15:21:54 | | | |
| 2 | Conner Chen | Meta information modification | 4269 | 2021-08-05 14:32:47 | | |
Video Upload Options
Heavy metals are one of the major pollutants that contribute to the escalating problem of environmental pollution, being primarily introduced in sensitive ecological habitats through industrial effluents, wastewater, as well as sewage of various industries. Microbial bioremediation, particularly the use of bacteria, has gained attention due to the feasibility and efficiency of using them in removing heavy metals from contaminated environments. Bacteria have several methods of processing heavy metals through general resistance mechanisms, biosorption, adsorption, and efflux mechanisms. Bacillus spp. are model Gram-positive bacteria that have been studied extensively for their biosorption abilities and molecular mechanisms that enable their survival as well as their ability to remove and detoxify heavy metals.
1. Bioremediation—An Environmentally-Friendly Approach for the Removal of Heavy Metals
Bioremediation is a conventional process that employs the use of plants, animals, and microorganisms for the remediation of pollutants like heavy metals. It is one of most effective, non-invasive, and economically feasible methods for the permanent mitigation of heavy metal pollution and for the complete restoration of the natural environment of many ecosystems, as there is no production of secondary by-products [1]. Though methods such as phytoremediation (bioremediation using plants), phycoremediation (bioremediation using algae) and mycoremediation (bioremediation using fungi) have been widely reported in literature for heavy metal removal, our review focuses on bacterial bioremediation with Bacillus spp. as potentially active and efficacious agents for the removal of various heavy metal ions present in the environment.
Bacteria are abundantly present in the environment, where their variety in shape, size, morphology, as well as resistance mechanisms makes them suitable for bioremediation of organic and inorganic pollutants. Biosorption mediated by bacteria is a cost-effective, economically feasible method for the removal of heavy metal ions and other pollutants from contaminated sites, as many species have evolved to develop mechanisms which mediate resistance to process heavy metal ions amidst high concentrations [2]. Over the past years, many bacterial species have garnered widespread attention for their potential ability to remove heavy metal ions from the environment, whereas bacterial biomass (live and dead) has also been investigated for biosorption of metal ions such as copper, chromium, zinc, nickel, cadmium, and mercury, concomitant on their interactions with the bacterial cell wall and related peptidoglycans [3][4][5]. Many bacterial species such as Bacillus, Pseudomonas, Enterobacter, Flavobacterium, Geobacter, and Micrococcus spp. have been investigated for potential use in biosorption of various heavy metal ions from contaminated sites, by observing their surface to volume ratio and other feasible characteristics which enhance the binding interactions between bacterial functional groups and heavy metal ions [6][7]. In a recent study, the biosorption ability of many bacterial species such as B. subtilis, B. megaterium, A. niger, and Penicillium spp. were investigated for the removal of many heavy metal ions such as lead, chromium, and cadmium, of which Bacillus spp. were observed to be the most efficient [8][9].
2. Bacillus Species and Heavy Metals
2.1. Arsenic
Arsenic (As) is a naturally occurring toxic metalloid [10] that is reported in soil (5–10 mg/kg), rock (1–2 mg/kg), sea water (1–3 µg/L) [11][12][13], as well as air and volcanic ash (0.02 µg/m3), due to which arsine gas (AsH3) and methylated arsine species enter into the surroundings [14]. Its anthropogenic sources are herbicides, pesticides [15], fossil fuel combustion, mining, smelting, wood preservation, sludge, manure [16], paint pigments, ceramic, glass industry, and food additives [17][18][19]. It exists as insoluble sulfides and sulfosalts [20]. It has four oxidation states: As3- (arsine), As0 (elemental arsenic), As3+ (arsenite), and As5+ arsenate. Among them, As3+ and As5+ are the most abundant forms. As3+ is more toxic and mobile in aqueous and oxic environments than As5+, which is mostly adsorbed to the sediments in an anoxic state [21][22]. The histidine part of cellular proteins is a target site for the interaction of thiols of cysteine residues and/or imidazolium nitrogen with As3+, which results in the inactivation of enzymes [23]. As5+ interferes with protein synthesis by replacing the phosphate group during phosphorylation of the energy transfer process. The existence of either state depends on the redox state of the environmental conditions [24], as well as the geomicrobial population [25]. The US EPA and WHO has allowed 10 µg/L as the permissible concentration of As in drinking water [16]. Above 0.5 ppm, it is toxic for living systems resulting in symptoms of skin cancer [26], loss of appetite, weakness, weight loss, lethargy, chronic respiratory disorders, gastrointestinal disorders, enlargement of liver, spleen disorders, anemia [27], and cardiovascular disease [28]. Introduction of arsenic into the food chain and ground water may lead to serious concerns of arsenicosis [22].
Bioregulation of Arsenic by Bacillus spp.
The soluble nature of As makes its removal from the environment difficult [29]. Physical and chemical remediation of arsenic involves an oxidation step for converting As3+ into As5+. It may occur under atmospheric oxygen, which is usually very slow, or by using chemical oxidants like hydrogen peroxide, chlorine, or ozone. This method is very expensive and produces harmful byproducts [30]. Microorganisms use As as an energy source in their metabolic processes and thus transform the toxic As3+ into its less toxic form, As5+ [31][32] by arsenic oxidase, which is present in the protoplasm of arsenic-oxidizing bacteria [10]. Liao et al. [33] reported Bacillus as one the important arsenic-reducing bacteria. Arsenic removal by Bacillus spp. like B. megaterium [34][35], B. aryabhattai [36], and B. cereus strain W2 was studied by Miyatake and Hayashi [37]. Anaerobic respiration of As5+ by B. cereus was reported by Ghosh et al. [22]. Various other studies conducted over the years have reported the ability of Bacillus spp. to uptake As [38][39][40][41][42] (Table 1). As speciation, solubility, and mobilization are dependent on its methylation [43][44], oxidation [45], reduction, and respiration. The significance of oxidative phosphorylation in living organisms cannot be ignored [19]. Structurally, As5+ has similarity with phosphate [16]; thus, it is a potential oxidative phosphorylation inhibitor [19]. As5+ enters the organism’s living system by employing two pathways; Pit and Pst [45], which are usually used for phosphate uptake. It interferes with phosphorylation metabolic reactions and inhibits synthesis of adenosine triphosphate [10]. The portal of entry for As3+ is aquaglyceroporin proteins [16][46][47]. After internalization, it immediately binds to the respiratory enzymes via their sulfur residue [17][48]. Bioremediation of As in Bacillus spp. is done by ars operon [49] (Table 2) by employing three genes arsA, arsB, arsC, arsD, and arsR. arsA and arsB have ATPase activity, arsC transforms As3+ to As5+, arsD works as metallochaperone, while arsR acts as a repressor [50][51][52][53]. Normally, As5+ (less toxic form) that enters the cell is reduced to As3+ (more toxic form) by ArsC and then transported out of the cell by ArsB [54][55] (Figure 1).
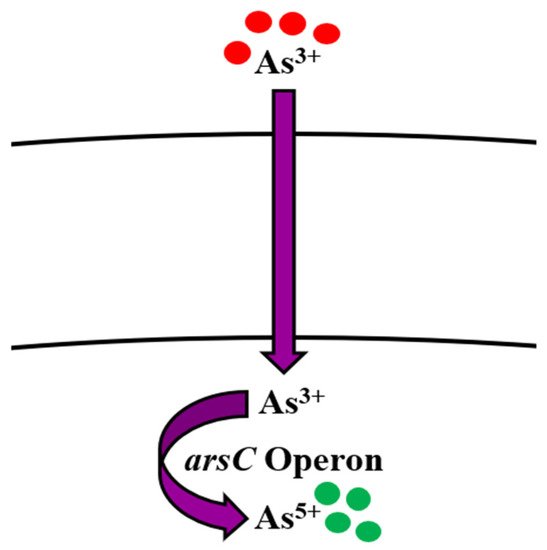
Figure 1. Bioregulation mechanism of arsenic in Bacillus spp.
Table 1. Uptake ability of Bacillus spp. for various heavy metals.
| Metal | Bacterial Strains | Initial Metal Concentration (%/mg/g/mM/ppm/mg/L) |
Metal Uptake Ability (%/mg/g/mM/ppm/mg/L/mol/g) |
Reference |
|---|---|---|---|---|
| Arsenic | Bacillus sp. KM02 | 100 ppm | 51.45% (As3+) | [10] |
| B. licheniformis B. polimyxa |
0–100 mM 0–20 mM |
100 ppm (As0) 100 ppm (As0) |
[39] | |
| Bacillus sp. IIIJ3–1 | 350 smM (As5+) 10 mM (As3+) |
350 mM (As5+) 10 mM (As3+) |
[22] | |
| B. barbaricus | - | 20 mM (As5+) 0.3 mM (As3+) |
[42] | |
| B. indicus Sd/3T | 0 mM 0 mM |
20 mM (As5+) 30 mM (As3+) |
[40] | |
| B. selenatiredreducens | 10 mM | 0 mM (As5+) 0.3 mM (As3+) |
[38] | |
| B. arsenicus con a/3 | 20 mM 0.5 mM |
20 mM (As5+) 0.3 mM (As3+) |
[41] | |
| B. cereus W2 | 50 mg/L | 1.870 mg/L (As3+) | [37] | |
| B. cereus EA5 B. fusiformis EA2 |
15 mg/L | 94.9% 99.7% |
[35] | |
| B. arsenicus MTCC 4380 | 2000 mg/L 1800 mg/L |
89.462% (As5+) 83.043% (As3+) |
[43] | |
| Zinc | B. subtilis | 178 mg/L | 49.7 mg/L | [56] |
| Bacillus sp. (KF710041) B. subtilis (KF710042) |
- | 73.29% 78.15% |
[57] | |
| B. licheniformis | - | 53% | [58] | |
| B. cereus | 0–200 mg/L | 66.6 mg/g | [59] | |
| B. jeotgali | 75 mg/l | 30% | [60] | |
| B. subtilis D215 | 100 mg/L | 63.73% | [61] | |
| B. firmus | 100 mg/L | 61.8% | [62] | |
| B. altitudinis | 100 mg/L | 87 mg/L | [63] | |
| Nickel | B. subtilis | 2.14 ppm | 85.61% | [64] |
| B. subtilis BM1 B. subtilis BM2 B. subtilis BM3 |
2–32 mg/L | 98.54% 99.2% 96.3% |
[65] | |
| B. subtilis | 178 mg/L | 57.8 mg/g | [56] | |
| Bacillus sp. KL1 | 100 ppm | 55.06% | [66] | |
| B. thuringiensis KUNi1 | 0–7.5 mM | 82% | [67] | |
| B. thuringiensis OSM29 | 25–150 mg/L | 94% | [68] | |
| B. thuringiensis | 250 mg/L | 15.7% | [69] | |
| Cadmium | B. safensis | 40 ppm 60 ppm |
83.5% 98.10% |
[70] |
| B. licheniformis | - | 98.34% | [71] | |
| B. catenulatus JB-022 | 150 mg/L | 66% | [72] | |
| B. thuringiensis DM55 | 0.25 mM | 79% | [73] | |
| Lead | B. pumilus MF472596 | 100–1000 ppm | 96% | [74] |
| B. subtilis X3 | 200–1400 mg/L | 590.49 mg/g | [75] | |
| B. cereus | 5–100 mg/L | 36.71 mg/g | [76] | |
| Bacillus S1 Bacillus SS19 |
75 and 100 mg/L 50 mg/mL |
53%, 51% 57% |
[77] | |
| Bacillus sp. AS2 | 500 ppm | 74.5 mg/g (99.5 %) | [78] | |
| Copper | B. cereus | 100 ppm | 54% | [79] |
| B. cereus | 400 ppm | 48% | [80] | |
| B. thuringiensis OSM29 | 25 mg/L | 91.8% | [68] | |
| B. licheniformis | 5 gm/L | 32% | [81] | |
| B. thioparans | 40 mg/L | 27.3 mg/g | [82] | |
| B. subtilis D215 | 100 mg/L | 67.18% | [61] | |
| B. sphaericus B. cereus Bacillus sp. |
17.6 mg/L 44.0 mg/L 88.0 mg/L |
5.6 mol/g 5.9 mol/g 6.4 mol/g |
[83] | |
| Bacillus sp. SG-1 | - | 60% | [84] | |
| Chromium | B. cereus NWUAB01 | 100 mg/L | 43% | [85] |
| B. cereus | 100 mg/L | 81% | [86] | |
| B. salmalaya 139SI | 50 ppm | 20.35 mg/g | [87] | |
| B. cereus FA-3 | 1000 μg/ml | 72% | [88] | |
| B. licheniformis | 15 mg/L | 95% | [89] | |
| Bacillus sp. B | 500–4500 mg/L | 47% | [90] | |
| B. marisflavi | 200 mg/L | 5.783% | [91] | |
| B. licheniformis | 300 mg/g | 69.4% | [92] | |
| B. thuringiensis | 250 mg/L | 83.3% | [93] | |
| B. licheniformis B. laterosporus |
- | 62 mg/g 72.6 mg/g |
[94] | |
| B. circulans B. megaterium |
0.96 mg/L | 34.5% 32% |
[95] | |
| Mercury | B. thuringiensis CASKS3 | 200 mg/L 400 mg/L 600 mg/L |
62.4% 54% 40% |
[96] |
| B. licheniformis | 50 mg/L | 70% | [97] | |
| B. cereus BW-03(pPW-05) | 5–50 ppm | 96.4% | [98] | |
| B. licheniformis | 100 μg/mL | 70% | [99] | |
| B. cereus | 5 mg/L | 104.1 mg/g | [100] | |
| Bacillus sp. | 1–10 mg/L | 7.9 mg/g | [101] | |
| Manganese | B. thuringiensis HM7 | 400 mg/L | 95.04% | [102] |
| B. cereus HM-5 | 600 mg/L | 67% | [103] | |
| Bacillus sp. | 13.3 mg/g | 55.56 mg/g | [104] | |
| Molybdenum | Bacillus sp. Zeid 14 | - | 200 mg/L | [105] |
| Bacillus sp. strain A.rzi | 0.1 mM | Not reported | [106] | |
| Silver | B. licheniformis R08 | 100 mg/L | 73.6 mg/g | [107] |
Table 2. The proteins, operons, and methods of removal employed by Bacillus spp. for the bioregulation of heavy metals.
| Metal | Protein(s)/Gene(s) | Method(s) | Reference |
|---|---|---|---|
| Arsenic | ars operon (arsR, arsD, arsA, arsB, arsC) | Reduction (Detoxification) Efflux Cell membrane binding, Adsorption on cell surface Complexation by exopolysaccharides |
[10][32][38][50][51][52][53] |
| Zinc | Zur ZosA ycdHI-yceA yciABC CadA CzcD |
Physico-chemical adsorption Ion exchange Efflux Uptake |
[59][108][109] |
| Nickel | CzcD CitM |
Efflux | [110][111] |
| Cadmium | cad operon yvgW KinA |
Efflux | [73][85][112][113][114][115][116] |
| Lead | pbr operon | Efflux | [117][118][119] |
| Copper | CueR copZA operon (CopA, CopZ, CopB) YcnJ |
Efflux by chaperone Uptake |
[120][121][122][123] |
| Chromium | ChrR | Efflux, Uptake Enzymatic reduction (Detoxification) |
[124][125][126] |
| Mercury | mer operon (merR, merA, merB) MerR MerA MerB |
Efflux Enzymatic reduction (Detoxification) |
[127][128][129][130][131] |
| Manganese | mntABCD operon MntR MntH MneP MneS |
Efflux Uptake |
[132][133][134][135] |
| Molybdenum | modABC operon | Uptake | [99][136][137][138][139][140][141] |
| Gold | Not reported | Bioaccumulation | [142][143] |
| Silver | SilP sil genes |
Efflux | [144][145][146][147][148][149] |
2.2. Zinc
Zinc (Zn) is one of the most profusely abundant transition elements in the Earth’s crust, and is also widely found in biological systems, coming second only to iron. Zn (atomic number 30) is a member of group XII (previously known as II-B) of the periodic table of elements, and as analogous to all members of the group, it is also characterized as a divalent metal [150]. At room temperature, it occurs as a brittle, lustrous metal with a blue-white hue [151]. In reactions concerning hydrolysis, Zn tends to act as a Lewis acid or electrophile, which catalyzes these reactions and is thereby integrated into assorted metallo-enzymes, transcription factors, and regulatory proteins [152]. In cells, it exhibits antioxidative properties against the formation and mitigation of free radicals and reactive oxygen species, which contributes to the perpetuation of protein stability. The significance of Zn resonates with its function as an essential nutrient in living systems, augmenting its presence in both human beings and bacteria, where more than 5% of bacterial proteins evince their dependency on Zn [153]. These manifold functions preponderate over its toxicity at higher concentrations in cells, which can often be promoted by the blockage of protein thiols via mis-metallation with other metals, resulting in the disruption of various biological functions [154].
In the environment, anthropogenic actions have shaped the presence of Zn and its compounds in industrial and agricultural wastewaters, underscoring the production (more than 12 million tons annually) and then consumption of Zn in a multitude of processes such as galvanization, metallurgy, and the pharmaceutical industry, alloy metal casting, pesticides, and production of several other consumer goods [155]. Moreover, mining activities and contamination of sludge in soils poses a considerable threat of Zn toxicity to the sustainability and quality of crops [156], which further raises concern for the purification of contaminated sites by effective methods. It has been reported that the removal of Zn in low concentrations is mediated by physical and chemical methods, but its removal by biological agents (plants, algae, microorganisms) is a method which has been gaining attention due to its many advantages that eclipse its drawbacks. The treatment of Zn-contaminated wastewaters through plants, biomass, sawdust, mollusk shells, fruit and vegetable peels, agricultural wastes, and polysaccharides such as chitosan and pectin as potentially effective biosorbents has been widely reported [157][158].
Bioregulation of Zinc by Bacillus spp.
The action of Bacillus spp. in removing Zn from contaminated environments has been highlighted in many bioremediation studies. This ability of Bacillus spp. is regulated either by acquiring resistance through plasmids or by evolving mechanisms of resistance [159][160]. In B. subtilis, Zn uptake is regulated by the Zur family, which enables the transport of Zn ions via two transporter proteins. Gaballa et al. [108] also reported a third uptake system in B. subtilis for Zn ions called ZosA (P-type ATPase), which was expressed in conditions of oxidative stress. Efflux of Zn in high concentrations is facilitated by a CPx-type ATPase efflux pump in B. subtilis, known as CadA [108][109] (Table 2, Figure 2). Moreover, the removal of Zn by Bacillus sp. such as B. subtilis, B. licheniformis, B. cereus, B. jeotgali, and B. firmus was reported in recent studies [56][57][58][59][60][61][62] (Table 1). Khan et al. [63] also reported the removal of Zn (87 mg/L) by B. altitudinis isolated from industrial wastewater.
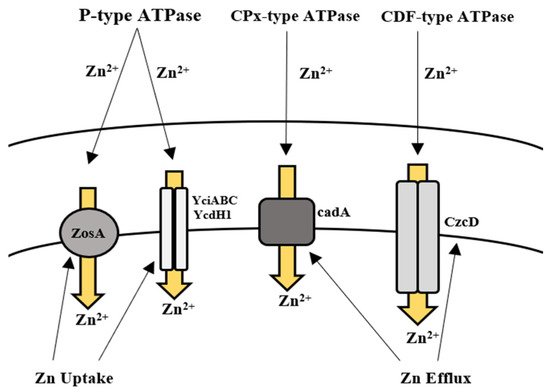
Figure 2. Uptake and efflux mechanism of Bacillus spp. for the regulation of zinc.
2.3. Nickel
Nickel (Ni) belongs to group 10 and is the 28th element in the periodic table, discovered by Swedish chemist Axel Cronstedt in its purified form for the first time in 1951. It is a hard, silvery-white transition metal which belongs to the ferromagnetic group of metals with high electrical and thermal conductivity [161]. It is the 24th most copious element found in the Earth’s crust, and the 5th most abundantly found in terms of weight. It is naturally found in its oxidation state (2+) which is analogous to most environmental and biological settings, though it may exhibit other valences as well (−1 to +4) [162]. It persists in nature in its hydroxide form at pH > 6.7, while its complexes appear to be readily soluble at pH < 6.5. It is found to exist in various forms of air- and water-resistant minerals (oxides and sulfides) [163], which give rise to Ni salts of strong (readily soluble in water) and weak acids (poorly soluble in water), respectively [164]. Natural sources of Ni in the environment are attributable to soil and rock erosion, volcanic eruptions, meteorite emissions, air-blown dust, as well as foods [165]. Combustion of fossil fuels and leaching from rocks and soil contribute to its presence in air and water, respectively. Moreover, anthropogenic emissions in the form of metal smelting and mining, metal refineries, Ni plating and alloy production, and effluent and sludge disposal into soil and water catalyze its presence in high concentrations in the environment [166]. The commercial use of Ni and its extensive applications such as production of Ni-Cd batteries, use in jewelry, orthodontic equipment, machinery, coins, food processing, clothing, and electronics promote its ubiquity in the environment, where it exists as sulfides, oxides, and less frequently, in its metallic form [167]. Ni toxicity has been the subject of widespread research in humans, where it is highlighted that the metal poses no considerable nutritional value in humans and poses industrial and occupational hazardous risk [168]. Nevertheless, it has been characterized as essential for the growth of plants, microorganisms and animals [169], where Ni-based enzymes and cofactors are reported to serve a key role in their function [170].
Bioregulation of Nickel by Bacillus spp.
There have been many methods of Ni removal from solid matrices, but the most effective are those which are capable of removing/treating Ni before it emanates into the environment [171]. Several physico-chemical methods have been employed over the years for the removal of Ni from aqueous solutions [172]. Regardless of which of these methods have been used in the past, newer, cheaper, and more efficient methods of adsorption have been used for Ni removal, such as the use of biomass, where sugarcane, corn cobs, citrus peels, and bark have been used [173]. In a study, corn hydrochar was treated with KOH and altered by treating polyethyleneimine (PEI) to increase adsorption of Ni ions onto the surface [174]. Bioremediation by Gram-positive bacteria, such as Bacillus spp., has been the better method for the removal of Ni ions from Ni-contaminated media. There have been many studies that demonstrate the uptake and/or removal of Ni ions from contaminated environments such as soils, wastewater, and rivers [56][64][65][66] (Table 1). B. thuringiensis has also been frequently reported to uptake and remove Ni from contaminated environments [67][68][69]. In a recent study, B. megaterium was isolated from Ni-contaminated soils and was able to uptake more than 500 mg Ni, where more than 3000 mg/L Ni salt was previously found [175]. The removal of Ni by bacteria is contingent on their inherent mechanisms of resistance, which ultimately facilitate uptake, transportation, and efflux of the metal ions in and out of the cell. According to Moore et al. [110], mechanisms of Ni homeostasis and regulation have not been well characterized in B. subtilis when compared to Gram-negative bacteria such as Escherichia coli and Helicobacter pylori, though some evidence suggests their presence [176]. Members of the cation diffusion facilitator (CDF) family have long been characterized to mediate efflux of multiple metal ions, including Ni [111]. In B. subtilis, the cation diffusion transporter CzcD is reported to provide protection to the cell amid high concentrations of Ni2+, Cu+, Zn2+, and Co2+ [110] (Figure 3). When these ions happen to be bound with citrate, these complexes, during favorable conditions, are taken up by the metal-dicitrate uptake system known as CitM in B. subtilis, consequently leading to an increase in toxicity to them [110] (Table 2).
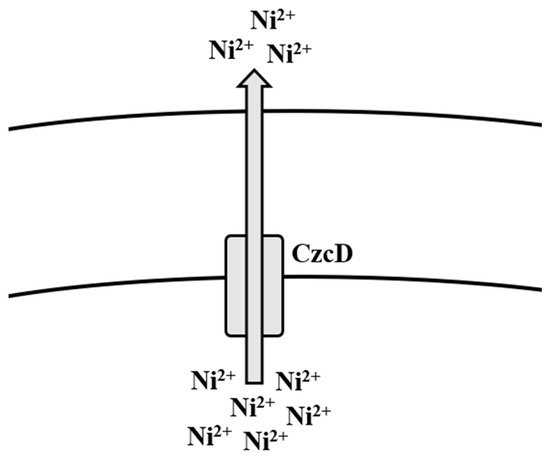
Figure 3. Efflux mechanism of nickel in Bacillus spp.
2.4. Cadmium
Cadmium (Cd) is a member of group XII of the periodic table and is a silvery white metal in appearance, with physical and chemical properties similar to both zinc and mercury. It exists in general oxidation state (+2) and is malleable and ductile. Cd is a corrosion-resistant metal, which is not flammable and water-soluble in nature, and is generally regarded as a toxic heavy metal with wide application in the industries of batteries, plating, plastics, and pigments, contributing greatly to its toxicity [170]. Anthropogenic activities have led to its presence being observed in many food sources and drinks [177]. Cadmium oxide is often used in metal plating, catalysis, and ceramic glaze. Alongside cadmium telluride, it has been also used in the form of a thin film for use in diodes, transistors, solar cells, electrodes, and anti-reflective coatings [178]. Cd is also used extensively in industrial strength paints, which can pose an environmental hazard during spraying. Other sources of contamination such as Cd-containing fertilizers can pollute soils which can inadvertently enhance its absorption by humans and other living beings. Cd has no known biological function and is a great threat to all life forms [179], due to which its environmental exposure can be dangerous, and in some cases, fatal [180].
Bioregulation of Cadmium by Bacillus spp.
Nature has gifted microorganisms with cadA operon to combat Cd toxicity. It is a 3.5 kb operon located on plasmid pI258. It has two genes; cadA and cadC [181][182]. cadA is transcribed into the 727-amino acid protein, which performs the function of energy-dependent Cd efflux ATPase [85], whereas cadC encodes relatively a small protein of 122 amino acids, and is a positive transcription regulator of Cd2+ operon [182][183]. It is well reported that cadA gene is induced in the presence of Cd2+ ions. Solovieva and Entian [112] documented their findings on cadA as a chromosomal determinant as well as a new gene yvgW of B. subtilis involved in Cd2+ resistance. Moreover, they reported that it has similarity to cadA of S. aureus plasmid pI258 [184]. Deletion of yvgW increased the Cd2+ sensitivity in the bacterium. cadB is also plasmid-mediated and confers Cd2+ resistance through a change in the binding site [113]. In addition to efflux mechanism, KinA and histidine kinase provide phosphate for phosphorylation which leads directly to transcription in B. subtilis [114]. Its overexpression results in phosphate flux of the cell, thereby directly affecting the energy state of the cell wall [185]. It also plays a significant role in biosorption of Cd2+ ions by phosphorylating the Bacillus cell surface magnitude, hence it acting as a major phosphate provider [73]. Cd2+ enters the bacterial cell membrane via the zinc and manganese transport systems, which are chromosomally mediated. Cd2+ concentration greatly affects the whole process. If the Cd2+ ions are present in high quantity in the medium, much extracellular adsorption is observed. Otherwise, intracellular Cd2+ concentration is high [186]. The cad operon of Bacillus megaterium TWSL_4 contains cadC, which has Cd2+ and Zn2+ metal binding motifs [187]. On exposure to metal ions, the first interaction is always with cell wall [188]. Its structure and composition play a significant role in deciding the next step of the process. Cd2+ adsorption on the bacterial cell wall deals with exchanging ions like Ca2+, Mg2+, and H+ ions [189]. Chelation is another process in which Cd2+ ions are exchanged with cell surface protons like –SO3H, –COOH, and –NH [190]. It involves sequestration via intracellular metallothionein (MT) [191]. Inorganic deposition of Cd2+ in the cell wall or inside cells can take place through interaction with hydroxide, carbonate, sulfate, and phosphorus [192]. In addition to metal concentration, biosorption is dependent on cell wall composition and cell physiology [193]. In Gram-positive bacterial species, resistance to Cd2+ is achieved by cadA system that is plasmid-borne. Cd2+ enters the bacterial cell by the MIT (metal ion transporter) system [115][116] (Table 2). The genes for Cd2+ resistance are mostly plasmid-mediated. According to Chen et al. [194] they are found on R plasmid along with antibiotic resistance genes, e.g., in pathogens including K. pneumoniae, P. aeruginosa, and S. aureus. These genes are reported to be directly involved in the uptake of Cd2+ ions from the environment (Figure 4). Basha and Rajaganesh [71] reported B. licheniformis to be a good biosorbent for Cd2+, as it removed more than 98% of Cd2+. Other species such as B. catenulatus and B. safensis are also reported to be effective in removing Cd2+ [70][72] (Table 1).
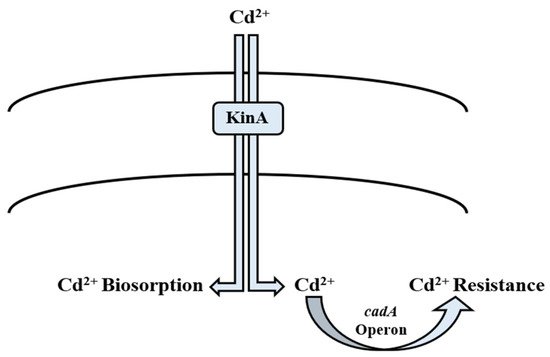
Figure 4. Mechanism of cadmium bioregulation by Bacillus spp.
2.5. Lead
Lead (Pb) is a toxic heavy metal that is introduced into the environment via the weathering of rocks. Anthropogenic sources include fossil fuels, extraction and melting of metals, battery-manufacturing industries, insecticides, pigments, and fertilizers [195]. Tetraethyl lead (TEL) has a common application as a gasoline additive, due to which it is a source of heat and electricity [196]. It exists in two states: Pb2+ and Pb4+ [197]. Its toxicity determines its bioavailability as well as mobility in the soil. Its common forms are oxides, hydroxides, ionic form, metal oxyanion complexes [151], phosphates, and carbonates (at pH above 6). The stable and insoluble forms include oxides, sulfides, and pyromorphites [198]. Its exposure occurs by food, water, and inhalation, which affects the circulatory, gastrointestinal, reproductive, neurological, muscular, kidney, and genetic systems [74]. Dose and exposure time are prime factors [199]. The permissible level of Pb in drinking water is <10 µL/L [182].
Bioregulation of Lead by Bacillus spp.
Pb-resistant Bacillus spp. have been reported previously [197]. Bacillus uses pbr operon [117][118][119] and active transport [200] as potential strategies to combat the toxic effects of Pb (Table 2, Figure 5). Microorganisms immobilize it by adsorption, chelation, inorganic precipitation, complexation, and biosorption. These processes involve bacterial cell wall functional groups including phosphate, carboxyl, carbonyl, sulfhydryl, and hydroxyl groups, which confer a negative charge to the cell wall. Binding of Pb to any of them results in insoluble substance. On the outside environment, Pb2+ is exchanged by Na or K cations [75]. Another method is adsorption through the cell wall, as it is comprised of organic macromolecules including polypeptides, polysaccharides, and proteins, which have the ability to adsorb Pb via electrostatic forces including Van der Waal’s forces, covalent or ionic bonds [75]. Pb interferes with microbial growth, morphology, and biochemical activities by damaging the DNA, protein, and lipids and even replacing the essential ions within the enzymes [74][201]. Microbes resist Pb toxicity by extracellular precipitation, exclusion, volatilization, biomethylation, cell surface binding, intracellular sequestration, and enhanced siderophore production [74]. Much like other Gram-positive bacteria, Bacillus spp. also employ one or several of these methods to remove Pb from the contaminated environments [76][77][78] (Table 1).
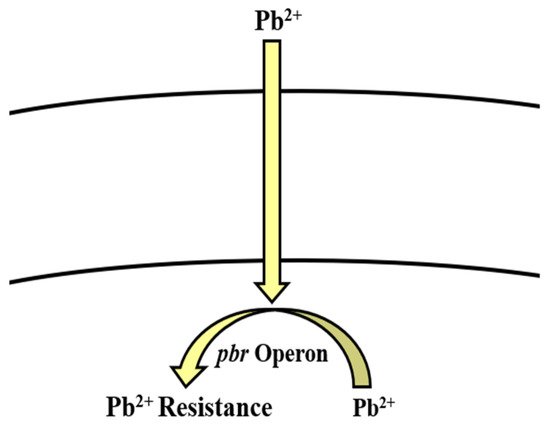
Figure 5. pbr operon involved in the regulation of lead resistance in Bacillus spp.
2.6. Copper
Copper (Cu) is categorized into the group I-B, and period 4 of the periodic table [151]. It is a soft, diamagnetic, malleable, and ductile metal with remarkable electrical and thermal conductivity. It acts as a soft and intermediate Lewis acid and tends to bind to soft bases (hydride, alkyl, thiol, phosphine) and auxiliary ligands to Cu2+, such as sulfate and nitrate [202]. Apart from being widespread in the environment thanks to anthropogenic actions, it also exists naturally in the form of minerals such as sulfides, carbonates, and oxides. The discovery and use of Cu dates back to ancient times, with its use spanning more than five thousand years. It exists in either of its two oxidation states, which can be the oxidized, divalent cupric form (Cu2+) or the reduced, monovalent cuprous form (Cu+) [203]. The significance of Cu in biological systems is pivotal; it serves an important role as a micronutrient in several biological processes in both prokaryotic and eukaryotic organisms. However, this stands only for lower concentrations of the metal; higher concentrations tend to induce cell toxicity, resulting in intracellular damage including changes in DNA, respiration, and overall growth [204]. Moreover, Cu is essentially required as a co-factor in more than thirty known enzymes, due to its ability to reversibly interconvert from its less to more required forms very easily [205]. Elevated levels of Cu exposure are a deep-rooted cause of environmental pollution by Cu, the fundamental reason being anthropogenic processes. Industries using Cu or its compounds, Cu mining, burning of fossil fuels, inadequate treatment of wastewater, accumulation in dumps, production of phosphate-containing fertilizer, and natural processes such as erosion, volcanic eruptions, forest wildfires, and decay are all processes which greatly contribute to its presence in the environment. Furthermore, its production is also a source of direct Cu pollution, capable of harming the fragile ecosystems of soil, water, and air, respectively [206].
Bioregulation of Copper by Bacillus spp.
Like many other heavy metal ions, Cu is considered to be essential for Bacillus subtilis, while concentrations exceeding normal amounts can be toxic for the cells. Species like B. thuringiensis, B. cereus, B. licheniformis, and B. sphaericus are also involved in the removal of Cu, when their concentrations exceed the required limit [68][79][80][81][82][83][84][207][208] (Table 1). In correlation with other bacterial species, Cu in the cytosol is regulated by CueR [209]. B. subtilis CueR is responsible for regulating the copZA operon which encodes both Cu chaperone and a P-type ATPase for Cu efflux, the latter of which is a member of the integral family which exports metal out of the cell [120]. In the former, CopZ plays a key role as Cu chaperone in transferring Cu over to CopA [121], which contributes to uptake of Cu, while CopB is accountable for Cu efflux and detoxification [122] (Table 2, Figure 1). Chillappagari et al. [123] reported that YcnJ was associated with Cu uptake function of copZA operon in B. subtilis, where their model proposed that the protein works in conjunction with other Cop proteins to facilitate Cu transport in and out of the cell (Figure 6).
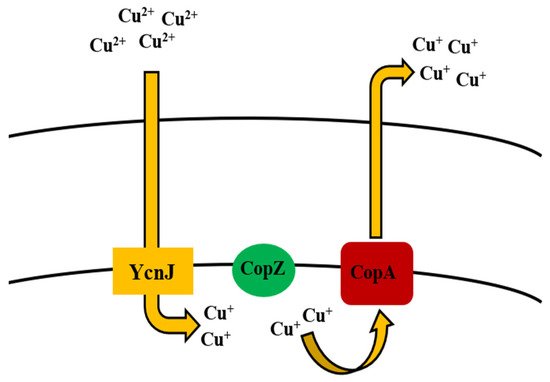
Figure 6. Uptake and efflux mechanism of copper by Bacillus spp.
References
- Abbas, S.H.; Ismail, I.M.; Mostafa, T.M.; Sulaymon, A.H. Biosorption of heavy metals: A review. J. Chem. Sci. Technol. 2014, 3, 74–102.
- Mustapha, M.U.; Halimoon, N. Microorganisms and biosorption of heavy metals in the environment: A review paper. J. Microb. Biochem. Technol. 2015, 7, 253–256.
- Kang, S.Y.; Lee, J.U.; Kim, K.M. Biosorption of Cr(III) and Cr(VI) onto the cell surface of Pseudomonas aeruginosa. Biochem. Eng. J. 2007, 36, 54–58.
- Akhtar, K.; Akhtar, M.W.; Khalid, A.M. Removal and recovery of uranium from aqueous solutions by Trichoderma harzianum. Water Res. 2007, 41, 1366–1378.
- Ozer, A.; Ozer, D. Comparative study of the biosorption of Pb(II), Ni(II) and Cr(VI) ions onto S. cerevisiae: Determination of biosorption heats. J. Hazard. Mater. 2003, 100, 219–229.
- Mosa, K.A.; Saadoun, I.; Kumar, K.; Helmy, M.; Dhankher, O.P. Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant. Sci. 2016, 7, 303.
- Kapahi, M.; Sachdeva, S. Bioremediation options for heavy metal pollution. J. Health Pollut. 2019, 9, 191203.
- Abioye, O.P.; Oyewole, O.A.; Oyeleke, S.B.; Adeyemi, M.O.; Orukotan, A.A. Biosorption of lead, chromium and cadmium in tannery effluent using indigenous microorganisms. Braz. J. Biol. Sci. 2018, 5, 25–32.
- Tarekegn, M.M.; Salilih, F.Z.; Ishetu, A.I. Microbes used as a tool for bioremediation of heavy metal from the environment. Cog. Food Agric. 2020, 6.
- Dey, U.; Chatterjee, S.; Mondal, N.K. Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotech. Rep. 2016, 10, 1–7.
- Matschullat, J. Arsenic in the geosphere—A review. Sci. Total Environ. 2000, 249, 297–312.
- WHO. Environmental Health Criteria: Arsenic and Arsenic Compounds; WHO: Geneva, Switzerland, 2001; p. 224.
- Oritz-Escobar, M.E.; Hue, N.V.; Cutler, W.G. Recent developments on arsenic: Contamination and remediation. In Recent Research Developments in Bioenergetics; Transworld Research Network: Trivandrum, India, 2006; Volume 4, pp. 1–32.
- Wilcox, D.E. Arsenic. Can This Toxic Metalloid Sustain Life? In Interrelations between Essential Metal Ions and Human Diseases. Metal Ions in Life Sciences; Sigel, A., Sigel, H., Sigel, R., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 475–498.
- Mandal, B.K.; Suzuki, K.T. Arsenic around the world: A review. Talanta 2002, 58, 201–235.
- Hue, N.V. Bioremediation of Arsenic Toxicity. In Arsenic Toxicity: Prevention and Treatment; Chakrabarty, N., Ed.; CRC Press: Boca Raton, FL, USA, 2015.
- Cervantes, C.; Ji, G.; Ramírez, J.L.; Silver, S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 1994, 15, 355–367.
- Ratnaike, R.N. Acute and chronic arsenic toxicity. Postgrad. Med. J. 2003, 79, 391–396.
- Lim, K.T.; Shukor, M.Y.; Wasoh, H. Physical, chemical, and biological methods for the removal of arsenic compounds. Biomed. Res. Int. 2014, 2014, 503784.
- Elangovan, D.; Chalakh, M.L. Arsenic pollution in West Bengal. Tech. Dig. 2006, 9, 31–35.
- Smedley, P.L.; Kinniburgh, D.G. A review of the source, behavior and distribution of arsenic in natural waters. Appl. Geochem. 2002, 17, 517–568.
- Ghosh, S.; Mohapatra, B.; Satyanrayana, T.; Sar, P. Molecular and taxonomic characterization of arsenic (As) transforming Bacillus sp. strain IIIJ3–1 isolated from As-contaminated groundwater of Brahmaputra river basin, India. BMC Microbiol. 2020, 20, 256–275.
- Mateos, L.M.; Ordonez, E.; Letek, M.; Gil, J.A. Corynebacterium glutamicum as a model bacteria for the bioremediation of arsenic. Int. Microbiol. 2006, 9, 2007–2015.
- Zhu, Y.G.; Yoshinaga, M.; Zhao, F.J.; Rosen, B.P. Earth abides arsenic biotransformations. Ann. Rev. Earth Planet. Sci. 2014, 42, 443–467.
- Paul, D.; Kazy, S.K.; Gupta, A.K.; Pal, T.; Sar, P. Diversity, metabolic properties and arsenic mobilization potential of indigenous bacteria in arsenic contaminated groundwater of West Bengal, India. PLoS ONE 2015, 19, e0118735.
- Khan, A.H.; Rasul, S.B.; Munir, A.; Habibuddowla, M.; Alauddin, M.; Newaz, S.S.; Hussan, A. Appraisal of a simple arsenic removal method for groundwater of Bangladesh. J. Environ. Sci. Health 2000, 35, 1021–1041.
- Banerjee, S.; Datta, S.; Chattyopadhyay, D.; Sarkar, P. Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J. Environ. Sci. Health Part A 2011, 46, 1736–1747.
- Smith, A.H.; Lingas, E.O.; Rahman, M. Contamination of drinking water by arsenic in Bangladesh: A public health emergency. Bull. World Health Org. 2000, 78, 1093–1103.
- Rhine, E.D.; Phelps, C.D.; Young, L.Y. Anaerobic arsenic oxidation by novel denitrifying isolates. Environ. Microbiol. 2006, 8, 899–908.
- Simeonova, D.D.; Micheva, K.; Muller, D.A.E.; Lagarde, F.; Lett, M.C.; Groudeva, V.I.; Lieremont, D. Arsenite oxidation in batch reactors with alginate-immobilized ULPAs1 strain. Biotechnol. Bioeng. 2005, 91, 441–446.
- Kamaluddin, S.P.; Arunkumar, K.R.; Avudainayagam, S.; Ramasamy, K. Bioremediation of chromium contaminated environments. IndianJ. Exp. Biol. 2003, 41, 972–985.
- Anyanwu, C.U.; Ugwu, C.E. Incidence of arsenic resistant bacteria isolated from a sewage treatment plant. Int. J. Basic Appl. Sci. 2010, 10, 64–78.
- Liao, V.H.-C.; Chu, Y.-J.; Su, Y.-C.; Hsiao, S.-Y.; Wei, C.-C.; Liu, C.-W.; Liao, C.-M.; Shen, W.-C.; Chang, F.-J. Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J. Contam. Hydrol. 2011, 123, 20–29.
- Miyatke, M.; Hayashi, S. Characteristics of arsenic removal from aqueous solution by Bacillus megaterium strain UM-123. J. Environ. Biotechnol. 2009, 9, 123–129.
- Mohamed, E.A.H.; Farag, A.G. Arsenic removal from aqueous solutions by different Bacillus and Lysinibacillus species. Bioremediat. J. 2015, 19, 269–276.
- Singh, N.; Gupta, S.; Marwa, N.; Pandey, V.; Verma, P.C.; Rathaur, S.; Singh, N. Arsenic mediated modifications in Bacillus aryabhattai and their biotechnological application for arsenic bioremediation. Chemosphere 2016, 164, 524–534.
- Miyatake, M.; Hayashi, S. Characteristics of arsenic removal by Bacillus cereus strain W2. Resour. Process. 2011, 58, 101–107.
- Blum, J.S.; Bindi, A.B.; Buzzelli, J.; Stolz, J.F.; Oremland, R.S. Bacillus arsenicoselenatis, sp. nov. and Bacillus selenitireducens sp. nov.: Two haloalkaliphiles from mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 1998, 171, 19–30.
- Anderson, C.R.; Cook, G.M. Isolation and characterization of arsenate-reducing bacteria from arsenic contaminated sites in New Zealand. Curr. Microbiol. 2004, 48, 341–347.
- Suresh, K.; Parabagaran, S.R.; Sengupta, S.; Shivaji, S. Bacillus indicus sp. nov. an arsenic-resistant bacterium isolated from an aquifer in West Bengal, India. Int. J. Syst. Evol. Microbiol. 2004, 54, 1369–1375.
- Shivaji, S.; Suresh, K.; Chaturvedi, P.; Dube, S.; Sengupta, S. Bacillus arsenicus sp. nov., an arsenic-resistant bacterium isolated from a siderite concretion in West Bengal, India. Int. J. Syst. Evol. Microbiol. 2005, 55, 1123–1127.
- Jiménez, G.; Blanch, A.R.; Tamames, J.; Rosselló-Mora, R. Complete genome sequence of Bacillus toyonensis BCT-7112T, the active ingredient of the feed additive preparation Toyocerin. Genome Announc. 2013, 1, 1–2.
- Podder, M.S.; Majumder, C.B. Biosorptive performance of Bacillus arsenicus MTCC 4380 biofilm supported on sawdust/MnFe2O4 composite for the removal of As(III) and As(V). Water Conser. Sci. Eng. 2016, 1, 103–125.
- Andreoni, V.; Zanchi, R.; Cavalca, L.; Corsini, A.; Romagnoli, C.; Canzi, E. Arsenite oxidation in Ancylobacter dichloromethanicus As3–1b strain: Detection of genes involved inn arsenite oxidation and CO2 fixation. Curr. Microbiol. 2012, 65, 212–218.
- Santini, J.M.; Stolz, J.F.; Macy, J.M. Isolation of a new arsenate-respiring bacterium-physiological and phylogenetic studies. Geomicrobiol. J. 2002, 19, 41–52.
- Rosen, B.P. Biochemistry of arsenic detoxification. FEBS Lett. 2002, 529, 86–92.
- Liu, Z.; Shen, J.; Carbrey, J.M.; Mukhopadhyay, R.; Agre, P.; Rosen, B.P. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP 9. Proc. Natl. Acad. Sci. USA 2002, 99, 6053–6058.
- Oremland, R.S.; Stolz, J.F. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005, 13, 45–49.
- Villegas-Torres, M.F.; Bedoya-Reina, O.C.; Salazar, C.; Vives-Florez, M.J.; Dussan, J. Horizontal arsC gene transfer among microorganisms isolated from arsenic polluted soil. Int. Biodeter. Biodegrad. 2011, 65, 147–152.
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212.
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Pathways of arsenic uptake and efflux. Curr. Top. Membr. 2012, 69, 325–358.
- Mukhopadhyay, R.; Rosen, B.P.; Phung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325.
- Zhou, T.; Radaev, S.; Rosen, B.P.; Gatti, D.L. Structure of the ArsA ATPase: The catalytic subunit of a heavy metal resistance pump. EMBO J. 2000, 19, 4838–4845.
- Afkar, E. Localization of the dissimilatory arsenate reductase in Sulfurospiriullum barnesii strain Ses-2. Am. J. Agric. Biol. Sci. 2012, 7, 97–105.
- Musingarimi, W.; Tuffin, M.; Cowan, D. Characterisation of the arsenic resistance genes in Bacillus sp. UWC isolated from maturing fly ash acid mine drainage Neutralised soilds. S. Afr. J. Sci. 2010, 106, 59–63.
- Wierzba, S. Biosorption of lead(II), zinc(II) and nickel(II) from industrial wastewater by Stenotrophomonas maltophilia and Bacillus subtilis. Pol. J. Chem. Technol. 2015, 17.
- Singh, P.P.; Chopra, A.K. Removal of Zn2+ and Pb2+ using new isolates of Bacillus spp. PPS03 and Bacillus subtilis PPS04 from Paper mill effluents using indigenously designed Bench-top Bioreactor. J. Appl. Nat. Sci. 2014, 6, 47–56.
- Kamika, I.; Momba, M.N. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 2013, 13, 28.
- Joo, J.H.; Hassan, S.H.; Oh, S.E. Comparative study of biosorption of Zn2+ by Pseudomonas aeruginosa and Bacillus cereus. Int. Biodeter. Biodegrad. 2010, 64, 734–741.
- Green-Ruiz, C.; Rodriguez-Tirado, V.; Gomez-Gil, B. Cadmium and zinc removal from aqueous solutions by Bacillus jeotgali: pH, salinity and temperature effects. Bioresour. Technol. 2008, 99, 3864–3870.
- Sabae, S.Z.; Hazaa, M.; Hallim, S.A.; Awny, N.M.; Daboor, S.M. Bioremediation of Zn+2, Cu+2 and Fe+2 using Bacillus subtilis D215 and Pseudomonas putida biovar A D225. Biosci. Res. 2006, 3, 189–204.
- Salehizadeh, H.; Shojaosadati, S.A. Removal of metal ions from aqueous solution by polysaccharide produced from Bacillus firmus. Water Res. 2003, 37, 4231–4235.
- Khan, M.; Ijaz, M.; Chotana, G.A.; Murtaza, G.; Malik, A.; Shamim, S. Bacillus altitudinis MT422188: A potential agent for zinc bioremediation. Bioremediat. J. 2021.
- Mardiyono; Sajidan; Masykuri, M.; Setyono, P. Bioremediation of nickel heavy metals in electroplating industrial liquid waste with Bacillus subtilis. AIP Conf. Proc. 2019, 2202.
- Al-Gheethi, A.; Mohamed, R.; Noman, E.; Ismail, N.; Kadir, O.A. Removal of heavy metal ions from aqueous solutions using Bacillus subtilis biomass pre-treated by supercritical carbon dioxide. Clean Soil Air Water 2017, 45, 1700356.
- Taran, M.; Sisakhtnezhad, S.; Azin, T. Biological removal of nickel (II) by Bacillus KL1 in different conditions: Optimization by Taguchi statistical approach. Pol. J. Chem. Technol. 2015, 17, 29–32.
- Das, P.; Sinha, S.; Mukherjee, S.K. Nickel Bioremediation potential of Bacillus thuringiensis KUNi1 and some environmental factors in nickel removal. Bioremediat. J. 2014, 18, 169–177.
- Oves, M.; Khan, M.S.; Zaidi, A. Biosorption of heavy metals by Bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi J. Biol. Sci. 2013, 20, 121–129.
- Öztürk, A. Removal of nickel from aqueous solution by the bacterium Bacillus thuringiensis. J. Hazard. Mater. 2007, 147, 518–523.
- Priyalaxmi, R.; Murugan, A.; Raja, P.; Raj, K.D. Bioremediation of cadmium by Bacillus safensis (JX126862), a marine bacterium isolated from mangrove sediments. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 326–335.
- Basha, S.A.; Rajaganesh, K. Microbial bioremediation of heavy metals from textile industry dye effluents using isolated bacterial strains. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 785–794.
- Kim, S.Y.; Jin, M.R.; Chung, C.H.; Yun, Y.-S.; Jahng, K.Y.; Yu, K.-Y. Biosorption of cationic basic dye and cadmium by the novel biosorbent Bacillus catenulatus JB-022 strain. J. Biosci. Bioeng. 2015, 119, 433–439.
- El-Helow, E.R.; Sabry, S.A.; Amer, R.M. Cadmium biosorption by a cadmium resistant strain of Bacillus thuringiensis: Regulation and optimization of cell surface affinity for metal cations. BioMetals 2000, 13, 273–280.
- Sahoo, S.; Goli, D. Bioremediation of lead by a halophilic bacteria Bacillus pumilus isolated from the mangrove regions of Karnataka. Int. J. Sci. Res. 2020, 9, 1337–1343.
- Qiao, W.; Zhang, Y.; Xia, H.; Luo, Y.; Liu, S.; Wang, S.; Wang, W. Bioimmobilization of lead by Bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. Open Sci. 2019, 6, 181701.
- Pan, J.-H.; Liu, R.-X.; Tang, H.-X. Surface reaction of Bacillus cereus biomass and its biosorption for lead and copper ions. J. Environ. Sci. 2007, 19, 403–408.
- Arifiyanto, A.; Apriyanti, F.D.; Purwaningsih, P.; Kalqutny, S.H.; Agustina, D.; Surtiningdih, T.; Shovitri, M.; Zulaika, E. Lead (Pb) bioaccumulation; genera Bacillus isolated S1 and SS19 as a case study. AIP Conf. Proc. 2017, 1854, 020003.
- Cephidian, A.; Makhdoumi, A.; Mashreghi, M.; Mahmudy Gharaie, M.H. Removal of anthropogenic lead pollutions by a potent Bacillus species AS2 isolated from geogenic contaminated site. Int. J. Environ. Sci. Technol. 2016, 13, 2135–2142.
- Raj, A.S.; Muthukumar, P.V.; Bharathiraja, B.; Priya, M. Comparative biosorption capacity of copper and chromium by Bacillus cereus. Int. J. Eng. Technol. 2018, 7, 442–444.
- Rohini, B.; Jayalakshmi, S. Bioremediation potential of Bacillus cereus against copper and other heavy metals. Int. J. Adv. Res. Biol. Sci. 2015, 2, 200–209.
- Karakagh, R.M.; Chorom, M.; Motamedi, H.; Kalkhajeh, Y.K.Y.; Oustan, S. Biosorption of Cd and Ni by inactivated bacteria isolated from agricultural soil treated with sewage sludge. Ecohydrol. Hydrobiol. 2012, 12, 191–198.
- Rodríguez-Tirado, V.; Green-Ruiz, C.; Gómez-Gil, B. Cu and Pb biosorption on Bacillus thioparans strain u3 in aqueous solution: Kinetic and equilibrium studies. Chem. Eng. J. 2012, 181, 352–359.
- da Costa, A.C.A.; Duta, F.P. Bioaccumulation of copper, zinc, cadmium and lead by Bacillus sp., Bacillus cereus, Bacillus sphaericus and Bacillus subtilis. Braz. J. Microbiol. 2001, 32.
- He, L.M.; Tebo, B.M. Surface charge properties of and Cu(II) Adsorption by Spores of the Marine Bacillus sp. Strain SG-1. Appl. Environ. Microbiol. 1998, 64, 1123–1129.
- Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Sci. Rep. 2020, 10, 19660–19671.
- Nayak, A.K.; Panda, S.S.; Basu, A.; Dhal, N.K. Enhancement of toxic Cr (VI), Fe, and other heavy metals phytoremediation by the synergistic combination of native Bacillus cereus strain and Vetiveria zizanioides L. Int. J. Phytoremediat. 2018, 20, 682–691.
- Dadrasnia, A.; Wei, K.S.C.; Shahsavari, N.; Azirun, M.S.; Ismail, S. Biosorption potential of Bacillus salmalaya Strain 139SI for removal of Cr(VI) from aqueous solution. Int. J. Environ. Res. Public Health 2015, 12, 15321–15338.
- Singh, N.; Verma, T.; Gaur, R. Detoxification of hexavalent chromium by an indigenous facultative anaerobic Bacillus cereus strain isolated from tannery effluent. Afr. J. Biotechnol. 2013, 12, 1091–1103.
- Samarth, D.P.; Chandekar, C.J.; Bhadekar, R. Biosorption of heavy metals from aqueous solution using Bacillus licheniformis. Int. J. Pure Appl. Sci. Technol. 2012, 10, 12–19.
- Chaturvedi, M.K. Studies on chromate removal by chromium-resistant Bacillus sp. isolated from tannery effluent. J. Environ. Prot. 2011, 2, 76.
- Mishra, S.; Doble, M. Novel chromium tolerant microorganisms: Isolation, characterization and their biosorption capacity. Ecotoxicol. Environ. Saf. 2008, 71, 874–879.
- Zhou, M.; Liu, Y.; Zeng, G.; Li, X.; Xu, W.; Fan, T. Kinetic and equilibrium studies of Cr (VI) biosorption by dead Bacillus licheniformis biomass. World J. Microbiol. Biotechnol. 2007, 23, 43–48.
- Şahin, Y.; Öztürk, A. Biosorption of chromium (VI) ions from aqueous solution by the bacterium Bacillus thuringiensis. Process. Biochem. 2005, 40, 1895–1901.
- Zouboulis, A.; Loukidou, M.; Matis, K. Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process. Biochem. 2004, 39, 909–916.
- Srinath, T.; Verma, T.; Ramteke, P.; Garg, S. Chromium (VI) biosorption and bioaccumulation by chromate resistant bacteria. Chemosphere 2002, 48, 427–435.
- Saranya, K.; Sundaramanickam, A.; Shekhar, S.; Swaminathan, S. Biosorption of mercury by Bacillus thuringiensis (CASKS3) isolated from mangrove sediments of southeast coast India. Ind. J. Geo-Mar. Sci. 2019, 48, 143–150.
- Upadhyay, K.H.; Vaishnav, A.M.; Tipre, D.R.; Patel, B.C.; Dave, S.R. Kinetics and mechanisms of mercury biosorption by an exopolysaccharide producing marine isolate Bacillus licheniformis. 3 Biotech 2017, 7, 313.
- Dash, H.R.; Das, S. Bioremediation of inorganic mercury through volatilization and biosorption by transgenic Bacillus cereus BW-03(pPW-05). Int. Biodeter. Biodegrad. 2015, 103, 179–185.
- Muneer, B.; Iqbal, M.J.; Shakoori, F.R.; Shakoori, A.R. Tolerance and biosorption of mercury by microbial consortia: Potential use in bioremediation of wastewater. Pak. J. Zool. 2013, 45, 247–254.
- Sinha, A.; Khare, S.K. Mercury bioremediation by mercury accumulating Enterobacter sp. cells and its alginate immobilized application. Biodegradation 2012, 23, 25–34.
- Green-Ruiz, C. Mercury (II) removal from aqueous solutions by nonviable Bacillus sp. from a tropical estuary. Bioresour. Technol. 2006, 97, 1907–1911.
- Huang, H.; Zhao, Y.; Xu, Z.; Ding, Y.; Zhou, X.; Dong, M. A high Mn(II)-tolerance strain, Bacillus thuringiensis HM7, isolated from manganese ore and its biosorption characteristics. PeerJ 2020, 8, e8589.
- Zhenggang, X.; Yi, D.; Huimin, H.; Liang, W.; Yunlin, Z.; Guiyan, Y. Biosorption characteristics of Mn (II) by Bacillus cereus Strain HM-5 isolated from soil contaminated by manganese ore. Pol. J. Environ. Stud. 2019, 463–472.
- Hasan, H.A.; Abdullah, S.R.S.; Kofli, N.T.; Kamaruddin, S.K. Biosorption of manganese in drinking water by isolated bacteria. J. Appl. Sci. 2010, 10, 2653–2657.
- Adnan, A.S.M.; Abu Zeid, I.M.; Ahmad, S.A.; Halmi, M.I.E.; Abdullah, S.R.S.; Masdor, N.A.; Shukor, M.S.; Shukor, M.Y. A molybdenum-reducing Bacillus sp. strain Zeid 14 in soils from Sudan that could grow on amides and acetonitrile. Malays. J. Soil Sci. 2016, 20, 111–134.
- Othman, A.R.; Bakar, N.A.; Halmi, M.I.; Johari, W.L.; Ahmad, S.A.; Jirangon, H.; Syed, M.A.; Shukor, M.Y. Kinetics of molybdenum reduction to molybdenum blue by Bacillus sp. strain A.rzi. Biomed. Res. Int. 2013, 2013, 371058.
- Sun, D.; Li, X.; Zhang, G. Biosorption of Ag(I) from aqueous solution by Bacillus licheniformis strain R08. Appl. Mech. Mater. 2013, 295–298, 129–134.
- Gaballa, A.; Wang, T.; Ye, R.W.; Helmann, J.D. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 2002, 184, 6508–6514.
- Mikhaylina, A.; Ksibe, A.Z.; Scanlan, D.J.; Blindauer, C.A. Bacterial zinc uptake regulator proteins and their regulons. Biochem. Soc. Trans. 2018, 46, 983–1001.
- Moore, C.M.; Gaballa, A.; Hui, M.; Ye, R.W.; Helmann, J.D. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 2005, 57, 27–40.
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339.
- Solovieva, I.M.; Entian, K.-D. Investigation of the yvgW Bacillus subtilis chromosomal gene involved in Cd2+ ion resistance. FEMS Microbiol. Lett. 2002, 208, 105–109.
- Zhang, H.; Zhou, Y.; Bao, H.; Zhang, L.; Wang, R.; Zhou, X. Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol. Res. 2015, 172, 1–6.
- Hoch, J.A. Regulation of the phsophorelay and the initiation of sporulation in Bacillus subtilis. Ann. Rev. Microbiol. 1993, 47, 441–465.
- Herzberg, M.; Bauer, L.; Kirsten, A.; Nies, D.H. Interplay between seven secondary metal uptake system is required for full metal resistance of Cupriavidus metallidurans. Metallomics 2016, 8, 313–326.
- Yu, X.; Ding, Z.; Ji, Y.; Zhao, J.; Liu, X.; Tian, J.; Wu, N.; Fan, Y. An operon consisting of a P-type ATPase gene and a transcriptional regulator gene responsible for cadmium resistances in Bacillus vietamensis 151–6 and Bacillus marisflavi 151–25. BMC Microbiol. 2020, 20, 18–30.
- Hynninen, A.; Touze, T.; Pitkanen, L.; Lecreulx, D.M.; Virta, M. An efflux transporter PbrA and a phosphatase PbrB cooperate in lead-resistance mechanism in bacteria. Mol. Microbiol. 2009, 74, 384–394.
- Silver, S. Bacterial resistance to toxic metal ions—A review. Gene 1996, 179, 9–19.
- Silver, S.; Phung, L.T. A bacterial view of periodic table: Genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005, 32, 587–605.
- Gaballa, A.; Helmann, J.D. Bacillus subtilis CPx-type ATPases: Characterization of Cd, Zn, Co and Cu efflux systems. Biometals 2003, 16, 497–505.
- Smaldone, G.T.; Helmann, J.D. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 2007, 153, 4123–4128.
- Odermatt, A.; Suter, H.; Krapf, R.; Solioz, M. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann. N. Y. Acad. Sci. 1992, 671, 484–486.
- Chillappagari, S.; Miethke, M.; Trip, H.; Kuipers, O.P.; Marahiel, M.A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 2009, 191, 2362–2370.
- Dhal, B.; Thatoi, H.; Das, N.; Pandey, B.D. Reduction of hexavalent chromium by Bacillus sp. isolated from chromite mine soils and characterization of reduced product. J. Chem. Technol. Biotechnol. 2010, 85, 1471–1479.
- Cervantes, C.; Campos-Garcia, J.; Devars, S.; Gutierrez-Corona, F.; Loza-Tavera, H.; Torres-Guzman, J.C.; Moreno-Sanchez, R. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 2001, 25, 335–347.
- Aguilar-Barajas, E.; Paluscio, E.; Cervantes, C.; Rensing, C. Expression of chromate resistance genes from Shewanella sp. strain ANA-3 in Escherichia coli. FEMS Microbiol. Lett. 2008, 285, 97–100.
- Permina, E.A.; Kazakov, A.E.; Kalinina, O.V.; Gelfand, M.S. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006, 6, 49.
- Das, S.; Dash, H.R.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 2016, 100, 2967–2984.
- Bogdanova, E.S.; Bass, I.A.; Minakhin, L.S.; Petrova, M.A.; Mindlin, S.Z.; Volodin, A.A.; Kalyaeva, E.S.; Tiedje, J.M.; Hobman, J.L.; Brown, N.L.; et al. Horizontal spread of mer operons among Gram-positive bacteria in natural environments. Microbiology 1998, 144, 609–620.
- Narita, M.; Chiba, K.; Nishizawa, H.; Ishii, H.; Huang, C.-C.; Kawabata, Z.; Silver, S.; Endo, G. Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiol. Lett. 2003, 223, 73–82.
- Medina, J.A.C.; Farias, J.E.; Hernndez, A.C.; Martinez, R.G.; Valdes, S.S.; Silva, G.H.; Jones, G.H.; Campos-Guillen, J. Isolation and characterization of mercury resistant Bacillus sp. from soils with an extensive history as substrates for mercury Extraction in Mexico. Geomicrobiol. J. 2013, 30, 454–461.
- Guedon, E.; Helmann, J.D. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 2003, 48, 495–506.
- Fisher, S.; Buxbaum, L.; Toth, K.; Eisenstadt, E.; Silver, S. Regulation of manganese accumulation and exchange in Bacillus subtilis W23. J. Bacteriol. 1973, 113, 1373–1380.
- Glasfeld, A.; Guedon, E.; Helmann, J.D.; Brennan, R.G. Structure of the manganese bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 2003, 10, 652–657.
- Guedon, E.; Moore, C.M.; Que, Q.; Wang, T.; Ye, R.W.; Helmann, J.D. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol. Microbiol. 2003, 49, 1477–1491.
- Meaden, G.T. The general physical properties of manganese metal. Met. Rev. 1968, 13, 97–114.
- Langenhoff, A.A.M.; Bronwers-Ceiler, D.L.; Engelberting, J.H.L.; Quist, J.J.; Wolkenfelt, J.P.N.; Zehnder, A.J.B.; Schraa, G. Microbial reduction of manganese coupled to toluene oxidation. FEMS Microbiol. Ecol. 1997, 22, 119–127.
- Zhongxing, S.; Guimin, W.; Yonggen, J. Relationship between environmental manganese exposure and children’s neurological behavior. Shanghai J. Prev. Med. 2017, 29, 288–291.
- Zeng, X.-C.; Chao, S.-H.; Zhu, M.-L.; Fan, X.-T.; Jiang, Y.-X.; Cao, H.-B. The contents of five trace elements in Panaxnotoginseng and the associated health risk. China Environ. Sci. 2016, 36, 293.
- Merchant, A.T.; Spatafora, G.A. A role for the DtxR family of metalloregulators in gram-positive pathogenesis. Mol. Oral Microbiol. 2014, 29, 1–10.
- Huang, X.; Shin, J.H.; Pinochet-Barros, A.; Su, T.T.; Helmann, J.D. Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol. Microbiol. 2016, 103, 253–268.
- Reith, F.; Lengke, M.F.; Falconer, D.; Craw, D.; Southam, G. The geomicrobiology of gold. ISME J. 2007, 1, 567–584.
- Karamushka, V.I.; Ulberg, Z.R.; Gruzina, T.G.; Podolska, V.I.; Pertsov, N.V. Study of the role of surface structural components of microorganisms in heterocoagulation with colloidal gold particles. Prikl. Biokhim. Microbiol. 1987, 23, 697–702.
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341–353.
- Woods, E.J.; Cochrane, C.A.; Percival, S.L. Prevalence of silver resistance genes in bacteria isolated from human and horse wounds. Vet. Microbiol. 2009, 138, 325–329.
- Elkrewi, E.; Randall, C.P.; Ooi, N.; Cottell, J.L.; O’Neill, A.J. Cryptic silver resistance is prevalent and readily activated in certain Gram-negative pathogens. J. Antimicrob. Chemother. 2017, 72, 3043–3046.
- Loh, J.V.; Percival, S.L.; Woods, E.J.; Williams, N.J.; Cochrane, C.A. Silver resistance in MRSA isolated from wound and nasal sources in humans and animals. Int. Wound J. 2009, 6, 32–38.
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver resistance in Gram-negative bacteria: A dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046.
- Kędziora, A.; Wernecki, M.; Korzekwa, K.; Speruda, M.; Gerasymchuk, Y.; Łukowiak, A.; Bugla-Płoskońska, G. Consequences of long-term bacteria’s exposure to silver nanoformulations with different physicochemical properties. Int. J. Nanomed. 2020, 15, 199–213.
- Jensen, W.B. The place of Zinc, Cadmium, and Mercury in the Periodic Table. J. Chem. Ed. 2003, 80, 952–961.
- Wuana, R.A.; Okieimen, F.E. Heavy Metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. Int. Sch. Res. Not. 2011, 2011, 402647.
- Cerasi, M.; Ammendola, S.; Battistoni, A. Competition for zinc binding in the host-pathogen interaction. Front. Cell. Infect. Microbiol. 2013, 3, 108–121.
- Rahman, M.T.; Karim, M.M. Metallothionein: A potential link in the regulation of zinc in nutritional immunity. Biol. Trace Elem. Res. 2018, 182, 1–13.
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350.
- Zinicovscaia, I.; Yushin, N.; Grozdov, D.; Abdusamadzoda, D.; Safonov, A.; Rodlovskaya, E. Zinc-containing effluent treatment using Shewanella xiamenensis biofilm formed on zeolite. Materials 2021, 14, 1760.
- Jain, D.; Kour, R.; Bhojiya, A.A.; Meena, R.H.; Singh, A.; Mohanty, S.R.; Rajpurohit, D.; Ameta, K.D. Zinc tolerant plant growth promoting bacteria alleviates phytotoxic effects of zinc on maize through zinc immobilization. Sci. Rep. 2020, 10, 1385–1398.
- Zhou, C.; Gong, X.; Han, J.; Guo, R. Removal of Pb(II) and Zn(II) from aqueous solutions by raw crab shell: A comparative study. Water Environ. Res. 2016, 88, 374–383.
- Rodrigues, A.C.D.; do Amaral Sobrinho, N.M.B.; dos Santos, F.S.; dos Santos, A.M.; Pereira, A.C.C.; Lima, E.S.A. Biosorption of toxic metals by water lettuce (Pistia stratiotes) biomass. Water Air Soil Pollut. 2017, 228, 156.
- Silver, S. Plasmid-determined metal resistance mechanisms: Range and overview. Plasmid 1991, 27, 1–3.
- Krishna, M.P.; Varghese, R.; Babu, V.A.; Jyothy, S.; Hatha, A.A.M. Bioremediation of Zinc Using Bacillus sp. Isolated from Metal Contaminated Industrial Zone. In Prospects in Bioscience: Addressing the Issues; Sabu, A., Augustine, A., Eds.; Springer India: Delhi, India, 2013; pp. 11–18.
- Das, K.K.; Das, S.N.; Dhundasi, S.A. Nickel, its adverse health effects & oxidative stress. Ind. J. Med. Res. 2008, 128, 412–425.
- Valko, M.; Morris, H.; Cronin, M. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208.
- Schaumlöffel, D. Nickel species: Analysis and toxic effects. J. Trace Elem. Med. Biol. 2012, 26, 1–6.
- Das, K.; Reddy, R.; Bagoji, I.; Das, S.; Bagali, S.; Mullur, L.; Khodnapur, J.; Biradar, M. Primary concept of nickel toxicity—An overview. J. Basic Clin. Physiol. Pharm. 2019, 30, 141–152.
- Cempel, M.; Nikel, G. Nickel: A review of its sources and environmental toxicology. Pol. J. Environ. Stud. 2006, 15, 375–382.
- Beattie, H.; Keen, C.; Coldwell, M.; Tan, E.; Morton, J.; McAlinden, J.; Smith, P. The use of bio-monitoring to assess exposure in the electroplating industry. J. Expo. Sci. Environ. Epidemiol. 2017, 27, 47–55.
- International Agency for Research on Cancer (IARC). Nickel and Nickel Compounds Monograph; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; WHO Press: Geneva, Switzerland, 2017; pp. 169–218.
- Buxton, S.; Garman, E.; Heim, K.E.; Lyons-Darden, T.; Schlekat, C.E.; Taylor, M.D.; Oller, A.R. Concise review of nickel human health toxicology and ecotoxicology. Inorganics 2019, 7, 89.
- Boer, J.L.; Mulrooney, S.B.; Hausinger, R.P. Nickel-dependent metalloenzymes. Arch. Biochem. Biophys. 2014, 544, 142–152.
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human health and environmental toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679.
- Cagnetta, G.; Huang, J.; Yu, G. A mini-review on mechanochemical treatment of contaminated soil: From laboratory to large-scale. Crit. Rev. Environ. Sci. Technol. 2018, 48, 723–771.
- Moghbeli, M.R.; Khajeh, A.; Alikhani, M. Nanosilica reinforced ion-exchange polyHIPE type membrane for removal of nickel ions: Preparation, characterization and adsorption studies. Chem. Eng. J. 2017, 309, 552–562.
- Li, X.; Wang, Y.; Cui, X.; Lou, Z.; Shan, W.; Xiong, Y.; Fan, Y. Recovery of silver from nickel electrolyte using corn stalk-based sulfur-bearing adsorbent. Hydrometallurgy 2018, 176, 192–200.
- Vakili, M.; Rafatullah, M.; Yuan, J.; Zwain, H.M.; Mojiri, A.; Gholami, Z.; Gholami, F.; Wang, W.; Giwa, A.S.; Yu, Y.; et al. Nickel ion removal from aqueous solutions through the adsorption process: A review. Rev. Chem. Eng. 2020, 20190047.
- Njokua, K.L.; Akinyede, O.R.; Obidi, O.F. Microbial remediation of heavy metals contaminated media by Bacillus megaterium and Rhizopus stolonifer. Sci. Afr. 2020, 10, e00545.
- Krom, B.P.; Huttinga, H.; Warner, J.B.; Lolkema, J.S. Impact of the Mg2+-citrate transporter CitM on heavy metal toxicity in Bacillus subtilis. Arch. Microbiol. 2002, 178, 370–375.
- Sinicropi, M.S.; Amantea, D.; Caruso, A.; Saturnino, C. Chemical and biological properties of toxic metals and use of chelating agents for the pharmacological treatment of metal poisoning. Arch. Toxicol. 2010, 84, 501–520.
- Lokhande, B.; Patil, P.S.; Uplane, M.D. Studies on cadmium oxide sprayed thin films deposited through non-aqueous medium. Mater. Chem. Phys. 2004, 84, 238–242.
- Yuan, Z.; Luo, T.; Liu, X.; Hua, H.; Zhuang, Y.; Zhang, X.; Zhang, L.; Zhang, Y.; Xu, W.; Ren, R. Tracing anthropogenic cadmium emissions: From sources to pollution. Sci. Total Environ. 2019, 676, 87–96.
- Kumar, S.; Sharma, A. Cadmium toxicity: Effects on human reproduction and fertility. Rev. Environ. Health 2019, 34, 327–338.
- Parsons, C.; Lee, S.; Kathariou, S. Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other Gram-positive bacteria. Mol. Microbiol. 2020, 113, 560–569.
- Endo, G.; Silver, S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1995, 177, 4437–4441.
- Yoon, K.P.; Silver, S. A second gene in the Staphylococcus aureus cad A cadmium resistance determinant of plasmid pI258. J. Bacteriol. 1991, 173, 7636–7642.
- Silver, S.; Phung, L.T. Bacterial plasmid-mediated heavy metal resistances: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–897.
- Lang, W.K.; Glassey, K.; Archibald, A.R. Influence of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J. Bacteriol. 1982, 151, 367–375.
- Abbas, S.Z.; Rafatullah, M.; Hossain, K.; Ismail, N.; Tajarudin, H.A.; Khalil, H.P.S.A. A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 2017, 15, 243–262.
- Kumari, W.M.N.H.; Thiruchittampalam, S.; Weerasinghe, M.S.S.; Chandrasekharan, N.V.; Wijayarathna, C.D. Characterization of a Bacillus megaterium strain with metal bioremediation potential and in silico discovery of novel cadmium binding motifs in the regulator, CadC. Appl. Microbiol. Biotechnol. 2021, 105, 2573–2586.
- Peraferrer, C.; Martinez, M.; Poch, J.; Villaescusa, I. Toxicity of metal-ethylene diaminetetraacetic acid solution as a function of chemical speciation: An approach for toxicity assessment. Arch. Environ. Contam. Toxicol. 2012, 63, 484–494.
- Rukhsana, F.; Butterly, C.R.; Baldock, J.A.; Xu, J.M.; Tang, C. Model organic compounds differ in priming effects on alkalinity release in soils through carbon and nitrogen mineralization. Soil Biol. Biochem. 2012, 51, 35–43.
- Blanco, A. Immobilization of Nonviable Cyanobacteria and Their Use for Heavy Metal Adsorption from Water in Environmental Biotechnology and Cleaner Bioprocesses; Oluguin, E.J., Sanchez, E.J., Hernandez, E., Eds.; Taylor and Francis: Philadelphia, PA, USA, 2000; p. 135.
- Qin, W.; Zhao, J.; Yu, X.; Liu, Z.; Chu, Z.; Tian, J.; Wu, N. Improving cadmium resistance in Escherichia coli through continuous genome evolution. Front. Microbiol. 2019, 10, 278.
- Özdemir, S.; Kilinc, E.; Poli, A.; Nicolaus, B.; Guven, K. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii subsp. Decanicus and Geobacillus thermoleovorans sub. sp. Stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206.
- Li, J.; Liu, Y.-R.; Zhang, L.-M.; He, J.-Z. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manag. 2019, 237, 552–559.
- Chen, M.; Li, Y.; Zhang, L.; Wang, J.; Zhenf, C.; Zhang, X. Analysis of gene expression provides insights into the mechanism of cadmium tolerance in Acidthiobacillus ferrooxidans. Curr. Microbiol. 2015, 70, 290–297.
- Kushwaha, A.; Hans, N.; Kumar, S.; Rani, R. A critical review on speciation, mobilization and toxicity of lead in soil-microbe-plant system and bioremediation strategies. Ecotoxicol. Environ. Saf. 2018, 147, 1035–1045.
- Pacyna, E.G.; Pacyna, J.M.; Fudala, J.; Strzelecka-Jastrzab, E.; Hlawiczka, S.; Panasiuk, D.; Nitter, S.; Pregger, T.; Pfeiffer, H.; Friedrich, R. Current and future emissions of selected heavy metals to the atmosphere from anthropogenic sources in Europe. Atmos. Environ. 2007, 41, 8557–8566.
- Varghese, R.K.M.P.; Arun, B.V.; Hatha, M.A.A. Biological removal of lead by Bacillus sp. obtained from metal contaminated industrial area. J. Microbiol. Biotechnol. 2012, 2, 756–770.
- Lindsay, W.L. Chemical Equilibria in Soils; John Wiley and Sons: Chichester, UK, 1979.
- Rigoletto, M.; Calza, P.; Gaggero, E.; Malandrino, M.; Fabbri, D. Bioremediation methods for the recovery of lead-contaminated soils: A review. Appl. Sci. 2020, 10, 3528.
- Silver, S.; Ji, G. Newer systems for bacterial resistances to toxic heavy metals. Environ. Health Perspect. 1994, 102, 107–113.
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Burkle, A. Interference by toxic metal ions with DNA repair processes and cell cycle control: Molecular mechanisms. Environ. Health Perspect. 2002, 110, 797–799.
- Crichton, R.R.; Pierre, J.L. Old iron, young copper: From Mars to Venus. BioMetals 2001, 14, 99–112.
- Osman, D.; Cavet, J.S. Copper Homeostasis in Bacteria. Adv. Appl. Microbiol. 2008, 65, 217–247.
- Zhao, S.L.; Liu, Q.; Qi, Y.T.; Duo, L. Responses of root growth and protective enzymes to copper stress in turfgrass. Acta Biol. Crac. Bot. 2010, 52, 7–11.
- Ma, Z.; Cowart, D.M.; Scott, R.A.; Giedroc, D.P. Molecular insights into the metal selectivity of the Cu(I)-sensing repressor CsoR from Bacillus Subtilis. Biochemisty 2009, 48, 3325–3334.
- Rehman, M.; Liu, L.; Wang, Q.; Saleem, M.H.; Ullah, B.; Peng, S. Copper environmental toxicology, recent advances, and future outlook: A review. Environ. Sci. Pollut. Res. 2019, 26, 18003–18016.
- Babak, L.; Šupinova, P.; Zichova, M.; Burdychova, R.; Vitova, E. Biosorption of Cu, Zn and Pb by thermophilic bacteria—Effect of biomass concentration on biosorption capacity. Acta Univ. Agric. Silvic. Mendel. Brunesis 2012, 60, 9–18.
- Liu, Y.-G.; Liao, T.; He, Z.-B.; Li, T.-T.; Wang, H.; Hu, X.-J.; Guo, Y.-M.; He, Y. Biosorption of copper(II) from aqueous solution by Bacillus subtilis cells immobilized into chitosan beads. Trans. Nonferr. Met. Soc. 2013, 23, 1804–1814.
- Changela, A.; Chen, K.; Xue, Y.; Holschen, J.; Outten, C.E.; O’Halloran, T.V.; Mondragon, A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 2003, 301, 1383–1387.




