
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Elisa Gamalero | + 2583 word(s) | 2583 | 2021-07-27 08:29:50 | | | |
| 2 | Nora Tang | + 29 word(s) | 2612 | 2021-07-30 10:29:04 | | | | |
| 3 | Nora Tang | + 29 word(s) | 2612 | 2021-07-30 10:29:22 | | |
Video Upload Options
While plant microbiomes may include bacteria, fungi, oomycetes and archaea, the majority of the studies published on this topic deal with bacteria (bacteriome), and to a lesser extent, fungi (mycobiome). Consequently, this review is directed toward developing an understanding of the functioning of bacteria within plant microbiomes. Although the microbiome of the phyllosphere impact plant health and, often, food production, only a limited number of studies have been aimed at discovering these particular communities, well adapted to the hostile leaf environment and mainly dominated by Alphaproteobacteria and by the generaMethylobacteriumandSphingomonas.
1. Introduction
An enormous number of bacteria are typically found in soil. Various soils contain ~1 × 106 to 1 × 109 bacterial cells per gram of soil, often including as many as 1 × 10 6 different taxa [1]. These bacteria may be beneficial (plant growth-promoting bacteria; PGPB), harmful (phytopathogens) or neutral in terms of their interaction with plants [2]. The greatest number of bacteria are typically found in the plant rhizosphere i.e., the region of the soil immediately around the roots [3]. The high concentration of bacteria occurring around plant roots is a direct consequence of the fact that plant roots commonly exude a significant fraction of the carbon that is fixed through photosynthesis [4][5]. In addition to being present in the rhizosphere, PGPB may also be endophytic, i.e., some bacteria are able to colonize the plant’s interior, symbiotic, i.e., some bacteria colonize the interior of the roots of specific plants by forming nodules on the plant root, or phyllospheric (i.e., they are found on the surfaces of plant leaves and stems) [2].
The vast majority of the reported laboratory and greenhouse studies of the interaction between soil bacteria and plants have been focused on the mechanisms used by individual bacterial strains, either PGPB or pathogens. However, in the past 10–15 years many scientists have turned their attention to the functioning of groups of bacteria, that often act together to affect plant growth and development. Much of the available evidence that exists to date indicates that different plants attract different cross-sections of the soil bacteria [1][6][7][8][9][10][11]. Each plant exudes or secretes through its roots a unique mixture of small molecules that attracts a specific fraction of the soil bacteria. In addition, different bacteria are attracted to and found in the plant microbiota (the characteristic microbial community occupying different parts of a plant). Thus, it is shown schematically in Figure 1 that microbiota that is found in a plant rhizosphere is quite different from the microbiota within the plant root tissues (the endosphere) and from the biota found in the bulk soil.
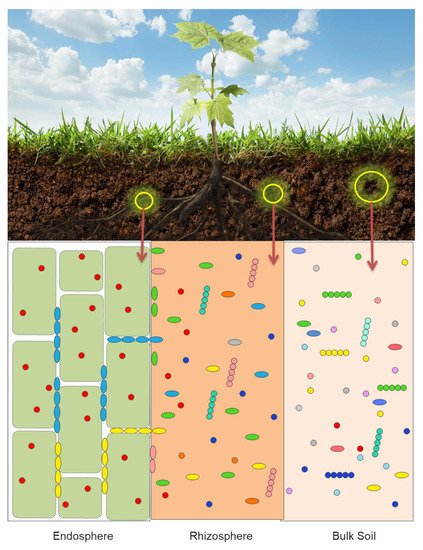
While plant microbiomes may include bacteria, fungi, oomycetes and archaea, the majority of the studies published on this topic deal with bacteria (bacteriome), and to a lesser extent, fungi (mycobiome). Consequently, this review is directed toward developing an understanding of the functioning of bacteria within plant microbiomes. Although the microbiome of the phyllosphere impact plant health and, often, food production, only a limited number of studies have been aimed at discovering these particular communities, well adapted to the hostile leaf environment and mainly dominated by Alphaproteobacteria and by the genera Methylobacterium and Sphingomonas [12]. Given that the plant root is the major site where plants and bacteria interact with one another, it is not surprising that the vast majority of soil bacteria are members of the root microbiota ( Figure 1 ). Root exudates are generally unique to each plant. As a consequence, different plants have a propensity for attracting specific subsets of soil bacteria. It is generally thought that the chemical composition of plant root exudates is involved in recruiting or selecting from the bulk soil the bacteria that make up a plant’s root microbiome [6][13][14][15][16][17]. However, the rhizosphere microbiota is only one of the key determinants of plant yield [18]. In addition to specific root exudates (a direct consequence of the plant host genotype), root microbiomes are also dependent upon soil type and environmental changes [19]. Oddly enough, different cultivars (subspecies) of the same plant can sometimes produce different panels of metabolites that make up their root exudates and therefore select for different root microbiomes. While plant-derived carbon appears to play a key role in determining the bacterial composition of the rhizosphere microbiome, soil-derived carbon compounds also can have an impact on the composition of rhizosphere microbiome [20]. Of the bacterial inhabitants of the plant rhizosphere, a small number of those bacteria are able to enter into the root tissues and subsequently permanently colonize the plant’s endosphere, i.e., they become the basis of an endophytic microbiome.
The soil and even the rhizosphere contain a wide variety of microorganisms including those that are beneficial and those that are potentially pathogenic. This being the case, it is essential to ask how plants recruit beneficial microorganisms, while for the most part, restrict (or try to restrict) pathogens. As indicated by Thoms et al. [21], “Once microbes are present, plants must decide to tolerate their presence, engage in mutualistic symbiosis, or mount an immune response”. These researchers have suggested that there are three stages in the interaction between plants and microorganisms. These include (i) metabolic gating where the production of specific metabolites by the plant restricts microbial access to the plant; (ii) dual receptor recognition in which specific signals from the plant and the microorganism bind to one another’s receptors and initiate either increased interaction or immunity; and (iii) integration of environmental signals with immune homeostasis. In metabolic gating, plants synthesize nutrients that only some microbes can use, or they can produce antimicrobial compounds that are toxic to only some microbes. Plant roots exude compounds using a variety of (mostly passive) mechanisms including diffusion, ion channels and vesicle transport. In general, ABC transporters are responsible for exudation of lipids and flavonoids, anion channels for sugars and other carbohydrates, metal transporters for various metals, aquaporins for water and uncharged molecules, and vesicles for high-molecular-weight compounds [4][5][22]. In dual receptor recognition, the exchange of signals combined with plant and microbial genetic potential helps to determine the ultimate fate of the interaction [23]. Finally, although microbes in the soil exist in the presence of a large number of other microbes, plants have to integrate their nutritional status with their expression of immunity to maintain homeostasis [21][24].
2. Soil Bacteria and Plant Growth
PGPB can benefit plant growth and development in a number of different ways and environmental conditions [2][25][26][27][28][29]. These include facilitating plant growth by increasing plant biomass, increasing plant mineral content (iron, phosphorus, potassium), providing plants with fixed nitrogen, increasing root and/or shoot length, enhancing seed germination, protecting plants from a wide range of phytopathogenic organisms (i.e., biocontrol), increasing plant tolerance to a wide range of environmental stresses (e.g., salt, flooding, drought, extremes of temperature, organic and inorganic soil contaminants), increasing the production of useful secondary metabolites, and increasing the overall level of plant nutrition. Conceptually, PGPB may facilitate plant growth both directly and indirectly [29]. Direct promotion of plant growth occurs when a PGPB either facilitates the acquisition of a required nutrient from the environment or adjusts and optimizes the level of hormones within a plant. As a consequence of the direct mechanisms, plants colonized by PGPB, show an increased plant biomass, yield and an improved nutritional value of seeds and fruits [30]. On the other hand, indirect promotion of plant growth reflects the ability of a PGPB to decrease the deleterious effects of phytopathogens on plants. Direct promotion of plant growth and development by PGPB may occur by producing auxin (most notably indoleacetic acid; IAA), ACC (1-aminocyclopropane-1-carboxylate) deaminase, cytokinin, or gibberellin, fixing atmospheric nitrogen, or solubilizing environmental phosphorus, iron or potassium. The indirect mechanisms used by PGPB to promote plant growth include ACC deaminase lowering of stress ethylene levels, synthesis of pathogen-inhibiting antibiotics, synthesis of pathogen fungal cell wall-degrading enzymes, outcompeting pathogens for nutrients (including for available iron) and/or space, synthesis of fungal pathogen-inhibiting hydrogen cyanide, and induction of systemic resistance mechanisms (ISR) [2][29] ( Figure 2 ).
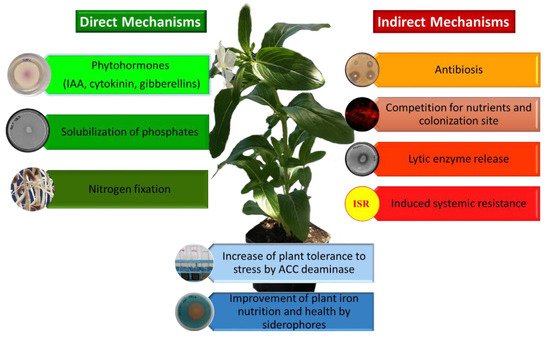
All these plant beneficial traits are at the base of the procedure for the selection of PGPB consisting in: (i) isolation of microorganisms from soil, rhizosphere or endosphere, (ii) characterization of the bacterial physiological activities, (iii) determination of the impact of the PGPB on the growth of plants under controlled and greenhouse conditions, and under optimal and stressed environmental conditions, (iv) assessment of the PGPB ecological safety, (v) development of a formulation satisfying the farmer’s needs and ensuring the bacterial viability, and (vi) marketing and registration. However, the occurrence of physiological plant beneficial strains is usually performed by qualitative or quantitative in-vitro tests, whose results do not always reflect the real PGPB performance in an open field [31]. Moreover, not all PGPB utilize the same mechanisms to facilitate plant growth; each encodes only a few of the above-mentioned mechanisms that are beneficial to plants. No one PGPB strain ever contains all of these traits. This is because increasing the number of genes that are involved in facilitating plant growth is likely to cause an increased metabolic load on the PGPB, thereby making it less competitive with other soil bacteria in the environment [32]. With this consideration, it is easy to understand why a microbiome containing a range of different PGPB (each with its own plant beneficial trait) might be more effective in promoting plant growth in the environment than a single (more limited) PGPB strain. Moreover, different plants may respond differently to a particular PGPB strain depending upon the phenological stage and the health status of the plant. Finally, it is important to keep in mind that the bacterial component is not unique in the rhizosphere. Other beneficial microorganisms such as mycorrhizal fungi, establishing mutualistic symbiosis with 90% of the land plants, strictly interact with the plant bacteriome, often leading to synergistic effects on plant growth (for a recent review see [33]).
3. Microbiomes and Stress
Various stresses can have a significant effect on plant metabolism and hence, on the composition of root exudates. Plants that are grown in the field are subjected to a wide range of both biotic and abiotic stresses. The biotic stress factors include pathogenic fungi, bacteria, viruses, nematodes and insects, all of which may be significantly deleterious to plant growth and health. In addition, a number of abiotic stresses may also negatively affect plant growth; these include high and low temperature, high light levels, drought, flooding, high salt, toxic organic compounds, inhibitory metals and radiation. Any one of these stresses can have a significant negative effect on plant growth and development. Moreover, a plant may sometimes encounter several environmental stresses at the same time. Of course, plants have their own built in defenses against many types of environmental stresses, however, in many cases a plant’s defenses provide insufficient protection against this environmental onslaught. Fortunately, a large number of rhizosphere microbiota protect plants against a wide range of environmental stresses. The protection against a wide range of environmental stresses provide a simple rationale for understanding why plants actively select PGPB. In exchange for the many ways in which PGPB facilitate plant growth, the bountiful root exudates provide PGPB with a much needed food source.
Despite the fact that different plants respond in various ways to biotic and abiotic stresses, nearly all environmental stresses induce plants to synthesize an increased level of the phytohormone ethylene [34]. Milder stresses cause the synthesis of low levels of ethylene that, in turn, induce the activation of the expression of plant defensive genes creating a protective response to the environmental stress in the plant. More severe or prolonged stresses often cause the synthesis of high levels of ethylene within the plant leading to plant senescence, chlorosis and abscission, typically exacerbating the effects of the environmental stress. Thus, when plants are highly stressed, “a large portion of the damage that occurs to the plant is due to autocatalytic ethylene synthesis” and not merely from the direct action of the stress [35]. A partial remedy to the deleterious and complicating effects of increased levels of ethylene is to treat plants (prior to the onset of any stress) with bacteria that synthesize the enzyme ACC deaminase [29][36][37][38][39][40][41][42]. In fact, lowering plant ethylene levels using PGPB that synthesize ACC deaminase, is a highly effective strategy to decrease the damage to plants caused by fungal pathogens, nematodes, flooding, drought, high salt, environmental contaminants and a number of other environmental stresses.
These limited number of experiments suggest that the presence of fungal pathogens (i.e., a biotic stress) alters the root exudates of plants and this change may favor the pathogenicity of the fungal pathogen. However, adding a suppressive compost, which is in effect adding a consortium of biocontrol bacteria, may overcome the negative outcome that would otherwise ensue from the presence of phytopathogens. It will be interesting to examine plant microbiomes following biotic stresses other than the presence of fungal pathogens to assess the nature of the changes to the plant microbiome.
The studies that have examined changes of the plant microbiomes as a consequence of abiotic stress, although mostly preliminary and incomplete, suggest that when plant growth is disturbed to a significant extent, the plant microbiome also is altered. The plant microbiome may, in some instances, protect the plant from the deleterious effects of abiotic stress [43]. Nevertheless, it is likely that different plant microbiomes will respond in different ways to the wide range and intensity of abiotic stresses.
4. Microbiomes of Transgenic Plants
Literature reports of studies of the microbiomes of transgenic plants are currently quite limited. However, this situation is likely to change in the next few years. More than 150 different plant species have already been genetically transformed and by 2018 more than 20 million farmers in 26 different countries worldwide were using this technology with major transgenic crops including soybean, corn, cotton, canola, alfalfa, rice, squash, papaya, wheat, eggplant, potatoes, sugarcane and apples [44]. While the biochemistry and physiology of a transgenic plant generally does not change to any appreciable extent, sometimes even different cultivars of the same plant have been observed to differ in the composition of their root exudates and rhizosphere/ endophere microbiota [45]. Thus, it is important to characterize the root exudates and microbiomes of transgenic plants to ensure that, where possible, we are able to understand and optimize the plant microbiomes of the transgenic plants to the same extent as their non-transgenic counterparts.
To date, scientists have reported the following. (i) When transgenic switchgrass plants that had been engineered to contain a decreased lignin content were grown in the field, over a period of two to five years, there was no effect of the transgenic plants on rhizospheric bacterial diversity, richness, or community composition [46]. (ii) In another study, rice plants were transformed with a Cry1Ab/1Ac gene which encodes a Bacillus thuringiensis insecticidal protoxin yielding a plant that is similar to other transgenic plants that have previously been released into the environment [47]. In this case, the transgenic rice did not confer any significant effect on the soil bacterial community structure. Again, this result suggests that generation of transgenic rice did not have a significant effect on the plant rhizosphere microbiome. (iii) A study of sugarcane plants that had been genetically engineered to overexpress the Ea-DREB2B gene in an effort to increase the drought tolerance of these plants revealed that the rhizosphere bacterial community of the transgenic plants were changed in response to changes in the plant root exudates [48]. From these limited studies, the preliminary conclusion may be drawn that transgenic plants do not have significantly altered root exudates and rhizosphere bacterial microbiomes unless the introduced transgene alters the behavior of the plant in the natural environment.
References
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838.
- Glick, B.R. Beneficial Plant-Bacterial Interactions, 2nd ed.; Springer: Heidelberg, Germany, 2020; p. 383.
- Lynch, J.M. The Rhizosphere; Wiley-Interscience: Chichester, UK, 1990; p. 458.
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in the rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266.
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant Physiol. 2003, 32, 44–51.
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424.
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41.
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37.
- Wang, N.R.; Haney, C.H. Harnessing the genetic potential of the plant microbiome. Biochemist 2020, 42, 20–25.
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621.
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690.
- Gong, T.; Xin, X.F. Phyllosphere microbiota: Community dynamics and its interaction with plant hosts. J. Integr. Plant Biol. 2021, 63, 297–304.
- Hartmann, A.; Schmid, M.; van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257.
- Chiellini, C.; Maida, I.; Emiliani, G.; Mengoni, A.; Mocoli, S.; Fabiani, A.; Biffi, S.; Maggini, V.; Gori, L.; Vannacci, A.; et al. Endophytic and rhizospheric bacterial communities isolated from the medicinal plants Echinacea purpurea and Echinacea angustifolia. Int. Microbiol. 2014, 17, 165–174.
- Antoniou, A.; Tsolakidou, M.-D.; Stringlis, I.A.; Pantelides, I.S. Rhizosphere microbiome recruited from a suppressive compost improves plant fitness and increases protection against vascular wilt pathogens of tomato. Front. Plant Sci. 2017, 8, 2022.
- Li, Y.; Wu, X.; Chen, T.; Wang, W.; Liu, G.; Zhang, W.; Li, S.; Wang, M.; Zhao, C.; Zhao, H.; et al. Plant phenotypic traits eventually shape its microbiota: A common garden test. Front. Microbiol. 2018, 9, 2479.
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Stanton, M.E. Soil indigenous microbiome and plant genotype cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201.
- Robertson-Albertyn, S.; Terrazas, R.A.; Balbirnie, K.; Blank, M.; Janiak, A.; Szarajko, I.; Chmielewska, B.; Karca, J.; Morris, J.; Hedley, P.E.; et al. Root hair mutation displace the barley rhizosphere microbiota. Front. Plant. Sci. 2017, 8, 1094.
- Kui, L.; Chen, B.; Chen, J.; Sharifi, R.; Dong, Y.; Zhang, Z.; Miao, J. A comparative analysis on the structure and function of the Panax notoginseng rhizosphere microbiome. Front. Microbiol. 2021, 12, 673512.
- Zhou, Y.; Yao, Q.; Zhu, H. Soil organic carbon attenuates the influence of plants on root-associated bacterial community. Front. Microbiol. 2020, 11, 594890.
- Thoms, D.; Liang, Y.; Haney, C.H. Maintaining symbiotic homeostasis: How do plant engage with beneficial microorganisms while at the same time restricting pathogens? Mol. Plant Microbe Interact. 2021.
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157.
- Zipfel, C.; Oldroyd, G.E.D. Plant signaling in symbiosis and immunity. Nature 2017, 543, 328–336.
- Singha, K.M.; Singh, B.; Pandey, P. Host specific endophytic microbiome diversity and associated functions in three varieties of scented black rice are dependent on growth stage. Sci. Rep. 2021, 11, 12259.
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401.
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39.
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197.
- Ali, S.; Glick, B.R. Plant-bacterial interactions in management of plant growth under abiotic stresses. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, J.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 21–45.
- Gamalero, E.; Glick, B.R. Plant growth-promoting bacteria in agriculture and stressed environments. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 361–380.
- Bona, E.; Lingua, G.; Todeschini, V. Effect of bioinoculants on the quality of crops. In Bioformulations: For Sustainable Agriculture; Arora, N., Mehnaz, S., Balestrini, R., Eds.; Springer: New Delhi, India, 2016.
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473.
- Glick, B.R. Metabolic load and heterologous gene expression. Biotechnol. Adv. 1995, 13, 247–261.
- Santoyo, G.; Gamalero, E.; Glick, B.R. Mycorrhizal-bacterial amelioration of plant abiotic and biotic stress. Front. Sustain. Food Syst. 2021, 5, 672881.
- Abeles, F.B.; Morgan, P.W.; Salveit, M.E., Jr. Ethylene in Plant Biology, 2nd ed.; Academic Press: New York, NY, USA, 1992; p. 41.
- Stearns, J.C.; Glick, B.R. Transgenic plants with altered ethylene biosynthesis or perception. Biotechnol. Adv. 2003, 21, 193–210.
- Glick, B.R. Phytoremediation: Synergistic use of plants and bacteria to clean up the environment. Biotechnol. Adv. 2003, 21, 383–393.
- Glick, B.R. Modifying a plant’s response to stress by decreasing ethylene production. In Phytoremediation and Rhizoremediation; Macková, M., Dowling, D.N., Macek, T., Eds.; Springer: Berlin, Germany, 2006; pp. 227–236.
- Glick, B.R.; Stearns, J.C. Making phytoremediation work better: Maximizing a plant’s growth potential in the midst of adversity. Int. J. Phytoremediat. 2011, 13, 4–16.
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633.
- Gamalero, E.; Glick, B.R. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015, 169, 13–22.
- Glick, B.R. Stress control and ACC deaminase. Principles of Plant-Microbe Interactions; Lugtenberg, B., Ed.; Springer Science: Berlin/Heidelberg, Germany, 2015; pp. 257–264.
- Gamalero, E.; Glick, B.R. The use of plant growth-promoting bacteria to prevent nematode damage to plants. Biology 2020, 9, 381.
- Timmusk, S.; Paalme, V.; Pavlicek, T.; Bergquist, J.; Vangala, A.; Danilas, T.; Nevo, E. Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 2011, 6, e17968.
- Glick, B.R.; Patten, C.L. Molecular Biotechnology: Principles and Applications of Recombinant DNA, 6th ed.; American Society for Microbiology: Washington, DC, USA, 2021; p. 1000. (in press)
- Chen, L.; Xin, X.; Zhang, J.; Redmile-Gordon, M.; Nie, G.; Wang, Q. Soil Characteristics overwhelm cultivar effects on the structure and assembly of root-associated microbiomes of modern maize. Pedosphere 2019, 29, 360–373.
- Debruyn, J.M.; Bevard, D.A.; Essington, M.E.; McKnight, J.Y.; Schaeffer, S.M.; Baxter, H.L.; Mazarei, M.; Mann, D.G.J.; Dixon, R.A.; Chen, F.; et al. Field-grown transgenic switchgrass (Panicum virgatum L.) with altered lignin does not affect soil chemistry, microbiology, and carbon storage potential. Glob. Chang. Biol. Bioenergies 2017, 9, 1100–1109.
- Wang, J.; Chapman, S.J.; Ye, Q.; Yao, H. Limited effect of planting transgenic rice on the soil microbiome studied by continuous 13CO2 labeling combined with high-throughput sequencing. Appl. Microbiol. Biotechnol. 2019, 103, 4217–4227.
- Zhao, X.; Jinag, Y.; Liu, Q.; Yang, H.; Wang, Z.; Zhang, M. Effects of drought-tolerant Ea-DREB2B transgenic sugarcane on bacterial communities in soil. Front. Microbiol. 2020, 11, 704.




