
Video Upload Options
Nanopore technology refers to nano-scale holes embedded in a thin membrane structure to detect the potential change when charged biological molecules smaller than nanopore pass through the hole. Therefore, nanopore technology has the potential to sense and analyze single-molecule amino acid, DNA, RNA, etc.. In this review, we will focus on the applications of nanopore technology in gene sequencing.
1. Introduction
Nanopore technology refers to nano-scale holes embedded in a thin membrane structure to detect the potential change when charged biological molecules smaller than nanopore pass through the hole [1]. Therefore, nanopore technology has the potential to sense and analyze single-molecule amino acid, DNA, RNA, etc. [2][3].
Nucleic acid is an important genetic material for most of the living body, and accurate sequencing of the nucleic acids is important for biomedical research, which would be useful for diagnosing human diseases and providing personalized medicine [4]. Since the last century, gene sequencing technology has developed dramatically, and now the nanopore technology has taken a leading role in the area of gene sequencing.
The development of gene sequencing technology. Since the 1950s, the determination of DNA double helix structure has preluded the research of genes. With the development of sequencing technology, the milestones have been made in gene sequencing. The gene sequencing technology has been constantly innovating and evolving in the direction of lower cost, higher throughput, and faster speed [5][6][7][8][9][10][11].
The first-generation sequencing technology is represented by Sanger sequencing, which mainly utilizes chain-terminating method and gel electrophoresis technology. However, the downside of the Sanger sequencing equipment was costly and time consuming. As the Human Genome Project cost $3 billion and took 13 years to complete [12], further application of first-generation gene sequencing technology was limited.
The second- and third-generation sequencing technologies are characterized by high throughput and are also known as Next Generation Sequencing Technologies (NGS) [13]. Roche’s 454 Sequencing Platform [14] and Illumina’s Solexa Sequencing System [15] are the representative. The BGISEQ-500 [16], a second-generation sequencing platform later developed by China’s BGI, has also taken some domestic market shares. Among the third-generation sequencing platforms, PacBio’s SMRT technology has become the backbone with its high throughput and high accuracy.
The fourth generation of gene sequencing technology was the combination between gene engineering technology and computer-aided technology. Oxford Nanopore sequencing technology (ONT) innovatively used the nanopore technology to determine the base sequences from the current distortion when DNA passing through the nanopore [17]. It has the advantages of the whole-genome sequencing and disease diagnosis with fast speed and cost-effective performance. 19].Table 1 shows the comparison of four kind sequencing devices of different sequencing generation.
| Reading Length (kb) N50 |
Estimated Cost per Gb (US $) | Throughput per Flow Cell (Gb) | Read Accuracy (%) | |
|---|---|---|---|---|
| Sanger(1st) | <1 kb | 13,000 d | / | >99.9 |
| Illumina(2nd) | 0.075–0.15 a | 50–63 | 16–30 | >99.9 |
| PacBio(3rd) | 10–20 b | 43–86 | 15–30 | >99 |
| ONT(4th) | 10–60 c | 21–42 | 50–100 | 87–98 |
There are several kinds of nanopore sequencing platforms, including ONT, NabSys, and Sequenom [21]. ONT uses biological nanopore to get the sequence of DNA or RNA. Different from ONT, NabSys uses solid-state nanopore to sequence and it can also reconstruct the DNA map by using the short DNA probe. As for nanopore sequencing technology of Sequenom, it uses a readout system technology for single DNA molecules based on simultaneous optical probing of multiple nanopore.
2. Principle of Nanopore Technology
Nanopore-based technologies originated from the Coulter and ion channels [22], which could be traced back to the 1990s [1]. Nanopore technology is done by applying a cathode and anode to the solution on the forward and reverse side of the membrane, respectively. Negatively charged biomolecules, such as DNA, can be placed on the forward side and these molecules can pass through pores in the membrane under the electrophoretic force with applied voltage. When different molecules translocate through the pores, the current level can be captured and be further used for calculation in computer-aided tools.
The category of nanopore used in nanopore technology can be divided into two parts, solid-state nanopore and biological nanopore. We first provide a brief overview of these nanopores in the following part.
Solid-state nanopore can be fabricated using different methods, such as controlled breakdown, electrochemical reactions, laser etching and laser-assisted controlled breakdown [23]. It breaks the limit of natural occurring nanopores and has many advantages, such as very high chemical stability, control of diameter and channel length, adjustable surface properties, and the potential for integration into devices and arrays [24][25].
Si3N4and SiO2nanopores are one of the most widely used nanopores and their fabrication is compatible with the complementary metal oxide semiconductor industrial integrated circuit processes [26]. These nanopores can be ion etched in free-standing Si3N4and SiO2films, using argon ion beam or electron beam [27].
Biological nanopore comes from natural protein molecules or artificial nanopores generated by genetic engineering [28]. However, the biological nanopores are frail with features such as short lifetime, intrinsic instability, and strict requirement of a specific environment, which are not able to support a biosensor’s long-term operations.
Biological nanopores are usually produced by selected bacteria, such as α-hemolysin pore protein [29], MspA from Mycobacterium smegmatis [30], and Phi29 from Bacillus subtilis [31]. These biological nanopores are currently used for disease diagnosis [32], gene sequencing [33], and protein sequencing [34].
3. Nanopore Sequencing Technology
ONT is a single molecule sequencing technology based on nanopores. The updated platform, PromethION, was released in 2015 with improved throughput. Two versions of PromethION, naming ProtheION 24 and 48, integrate 24 and 48 flow tanks, respectively. With the booming numbers of flow tanks compared to MinION, the PromethION system could output up to 7.6 Tb data while MinION could only generate 50 Gb within 72-h operation.
There are three forms of nanopore sequencing, 1D, 2D, and 1D2. After completing the sequencing of one strand, the sequencing of the other strand begins immediately. In this way, it is equivalent to repeat the sequencing twice, which can be used for base correction. 1D2is similar to 2D, whereas it doesn’t need hairpins to physically bind the two strands of DNA together.
The reaction system for nanopore sequencing is carried out in a flow cell, in which two ionic solution-filled compartments were separated by membranes containing either 2048 (MinION) or 12,000 (PromethION) nanopores. The process of nanopore gene sequencing can be divided into three parts, library preparation, sequencing process, and basecaller.
The preparation of the library is crucial for the subsequent work of nanopore sequencing. The DNA fragments should be repaired whether it has been sheared or not. When repairing, the repaired connector is a DNA-protein complex with a polymerase or helicase on the complex.
Figure 1 shows the schematic process of sequencing. When the DNA-protein complex approaches the nanopore, the enzyme binds to a single stranded leader at the end of the double stranded DNA template, unzips the double strand, and feeds a single strand through the nanopore. A single molecule with high specificity can interfere with the current when the unzipped DNA long strand passing through the nanopore one base at a time. These current signals can be used to determine the type of base.
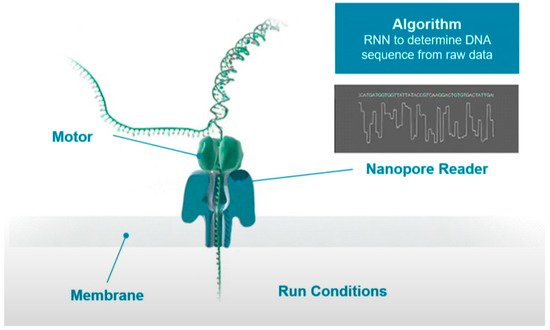
In the process of base readout, owing to the difference in the charge and structure of nucleotides when they translocate through the nanopore, the measured current would cause small disturbance. These electrical signals can then be translated into DNA sequences with the deep learning algorithms. However, the readout signals are noisy and random as these signals are originated from more than one molecule in the nanopores, which is difficult for the basecaller.
In addition, the resistance of the hole is determined by the bases of multiple nucleotides located at the narrowest point of the hole [36]. Being the last step for the interpretation of the entire DNA sequence, the data analysis using deep learning is challenging, which requires efficient algorithms and large amount of data for computational training.
References
- Deamer, D.W.; Akeson, M. Nanopores and nucleic acids: Prospects for ultrarapid sequencing. Trends Biotechnol. 2000, 18, 147–151.
- Rodriguez-Larrea, D. Single-aminoacid discrimination in proteins with homogeneous nanopore sensors and neural networks. Biosens. Bioelectron. 2021, 180, 7.
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.H.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153.
- Zhu, X.Q.; Li, J.; He, H.P.; Huang, M.; Zhang, X.H.; Wang, S.F. Application of nanomaterials in the bioanalytical detection of disease-related genes. Biosens. Bioelectron. 2015, 74, 113–133.
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467.
- Smith, L.; Sanders, J.; Kaiser, R.; Hughes, P.; Dodd, C.; Connell, C.; Heiner, C.; Kent, S.; Hood, L. Fluorescence detection in Automated DNA sequence analysis. Nature 1986, 321, 674–679.
- Connell, C.; Fung, S.; Heiner, C.; Bridgham, J.; Chakerian, V.; Heron, E.; Jones, B.; Menchen, S.; Mordan, W.; Raff, M.; et al. Automated DNA sequence analysis. BioTechniques 1987, 5, 342–348.
- Ronaghi, M.; Karamohamed, S.; Pettersson, B.; Uhlen, M.; Nyrén, P. Real-Time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996, 242, 84–89.
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380.
- McCarthy, A. Third Generation DNA sequencing: Pacific biosciences’ single molecule real time technology. Chem. Biol. 2010, 17, 675–676.
- Maxam, A.M.; Gilbert, W.J.B. A new method for sequencing DNA. 1977. Biotechnology 1992, 24, 99–103.
- Lander, E.S.E.A. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- Mardis, E.R. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402.
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769.
- Huang, W.; Li, L.; Myers, J.R.; Marth, G.T. ART: A next-generation sequencing read simulator. Bioinformatics 2012, 28, 593–594.
- Fang, C.; Zhong, H.; Lin, Y.; Chen, B.; Han, M.; Ren, H.; Lu, H.; Luber, J.M.; Xia, M.; Li, W.; et al. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 2018, 7, 1–8.
- Mikheyev, A.S.; Tin, M.M.Y. A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour. 2014, 14, 1097–1102.
- Patel, N.; Ferns, B.R.; Nastouli, E.; Kozlakidis, Z.; Kellam, P.; Morris, S. Cost analysis of standard Sanger sequencing versus next generation sequencing in the ICONIC study. Lancet 2016, 388.
- Ameur, A.; Kloosterman, W.P.; Hestand, M.S. Single-Molecule Sequencing: Towards Clinical Applications. Trends Biotechnol. 2019, 37, 72–85.
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21, 597–614.
- Sun, X.; Song, L.; Yang, W.; Zhang, L.; Liu, M.; Li, X.; Tian, G.; Wang, W. Nanopore sequencing and its clinical applications. Methods Mol. Biol. 2020, 2204, 13–32.
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based fourth-generation DNA sequencing technology. Genom. Proteom. Bioinform. 2015, 13, 4–16.
- Fried, J.P.; Swett, J.L.; Nadappuram, B.P.; Mol, J.A.; Edel, J.B.; Ivanov, A.P.; Yates, J.R. In situ solid-state nanopore fabrication. Chem. Soc. Rev. 2021, 50, 4974–4992.
- Ma, Q.; Si, Z.X.; Li, Y.; Wang, D.G.; Wu, X.L.; Gao, P.C.; Xia, F. Functional solid-state nanochannels for biochemical sensing. Trac. Trends Anal. Chem. 2019, 115, 174–186.
- Khan, M.S.; Dosoky, N.S.; Berdiev, B.K.; Williams, J.D. Electrochemical impedance spectroscopy for black lipid membranes fused with channel protein supported on solid-state nanopore. Eur. Biophys. J. Biophys. Lett. 2016, 45, 843–852.
- Tang, Z.F.; Zhang, D.H.; Cui, W.W.; Zhang, H.; Pang, W.; Duan, X.X. Fabrications, applications and challenges of solid-state nanopores: A mini review. Nanomater. Nanotechnol. 2016, 6, 12.
- Kim, M.J.; McNally, B.; Murata, K.; Meller, A. Characteristics of solid-state nanometre pores fabricated using a transmission electron microscope. Nanotechnology 2007, 18, 5.
- Mohammad, M.M.; Iyer, R.; Howard, K.R.; McPike, M.P.; Borer, P.N.; Movileanu, L. Engineering a rigid protein tunnel for biomolecular detection. J. Am. Chem. Soc. 2012, 134, 9521–9531.
- Bayley, H.; Cremer, P.S. Stochastic sensors inspired by biology. Nature 2001, 413, 226–230.
- Derrington, I.M.; Butler, T.Z.; Collins, M.D.; Manrao, E.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Nanopore DNA sequencing with MspA. Proc. Natl. Acad. Sci. USA 2010, 107, 16060–16065.
- Manrao, E.A.; Derrington, I.M.; Laszlo, A.H.; Langford, K.W.; Hopper, M.K.; Gillgren, N.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012, 30, 349–353.
- Brown, E.; Freimanis, G.; Shaw, A.E.; Horton, D.L.; Gubbins, S.; King, D. Characterising foot-and-mouth disease virus in clinical samples using nanopore sequencing. Front. Vet. Sci. 2021, 8, 10.
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Ettore, S.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016, 530, 228–232.
- Hu, Z.L.; Huo, M.Z.; Ying, Y.L.; Long, Y.T. Biological nanopore approach for single-molecule protein sequencing. Angew. Chem. Int. Edit. 2020, 133.
- Oxford Nanopore Technologies. Introduction to Real Time Analysis. 2020. Available online: https://www.youtube.com/watch?v=8oNEjt5Ov_Q (accessed on 2 April 2021).
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019.




