Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shuang Wang | + 1721 word(s) | 1721 | 2021-07-20 10:49:59 |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, S. DNA Assembly. Encyclopedia. Available online: https://encyclopedia.pub/entry/12513 (accessed on 08 February 2026).
Wang S. DNA Assembly. Encyclopedia. Available at: https://encyclopedia.pub/entry/12513. Accessed February 08, 2026.
Wang, Shuang. "DNA Assembly" Encyclopedia, https://encyclopedia.pub/entry/12513 (accessed February 08, 2026).
Wang, S. (2021, July 28). DNA Assembly. In Encyclopedia. https://encyclopedia.pub/entry/12513
Wang, Shuang. "DNA Assembly." Encyclopedia. Web. 28 July, 2021.
Copy Citation
Assembly of DNA-grafted 3D superlattice is composed of two pivotal elements: nanoparticles (NPs) core and DNA shell. The progress of chemical synthesis technique enables the chemical composition of NP core unrestricted; it can be metals, oxides, polymers, even biological molecule, which also allow for the integration of multiple functions like luminescent, catalytic, magnetic properties, and so on.
DNA nanotechnology
self-assembly
superlattice
DNA tile
DNA origami
1. Introduction
Self-assembly could arrange individual components into ordered structure, this phenomenon exists widely in nature, such as the double helix structure of DNA, protein aggregation and folding, cell, or some living life, and so on, they are considered to be the result of molecular self-assembly [1]. Inspired by the nature phenomenon and the unceasing understanding for self-assembly rules, researchers try to take atomic/molecular/ensembles in nanoscale as building blocks to fabricate devices, the automatic assembly among building blocks will obtain new structure/material with specific properties, which breakthrough the limitation of traditional top-down processing technology and meet the human needs in information, life, electronic, material fields. The assembly rules and methodologies are of great significance and dominate the material science field.
Colloid is often used as a model of atomic system to reveal the crystallization rules and the role of nanoparticle in assembly process [2]. Colloid nanoparticles as building blocks are linked together by weak interactions such as van der Waals, hydrogen bonding, or others to surface-bound ligands such as DNA [3], block copolymer [4], or small molecule [5]. Colloid particles protected by DNA with exceptional programmability and addressability exhibit tremendous potential in superlattice assembly. DNA-grafted nanoparticle as “programmable atom equivalent” (PAE) has been used in probing colloid assembly systems for many years.
2. Assembly Methods for 3D Lattice
2.1. Connection via ssDNA
The simplest but typical method for 3D lattice assembly is two NPs connected to lattice by the complementary pairing of single strand DNA. In 2008, Gang’s group displayed the effect of different (linker and spacer) DNA structures on the long-range order of NP lattice [6]. The DNA linker in the middle offers driving force for A and B NPs assembly (Figure 3A). The length of linker sequence and the free spacer were designed to tune the attraction and repulsive interaction among NPs, which guide the transition from disordered NPs to 3D ordered lattice. According to the experimental results, the longer spacers (system IV and V) are beneficial for the long-range order of crystalline organization, and the shorter or more rigid spacers (system I–III) showed amorphous upon annealing. Besides, the melting temperature (Tm) of DNA linker sequence is a turning point for crystallization. The formation of lattice is kinetically prohibited when the temperature is much below Tm due to the local DNA crowding and high hybridization energy. While the DNA attraction energy will be reduced when the system is heated to Tm, which leads to the uniaxial DNA hybridization and distance uniformity between NPs and achieves optimal crystalline packing. The DNA design produces profound effect on forming ordered 3D lattice via the equilibrium balance of repulsion and attraction of assembly system. The difference of assembly method in Mirkin’s group [7] is the introduction of another linker DNA (Figure 3B) which includes two regions. The longer region is complementary with the DNA decorated on NPs, and the shorter region was designed to control the interaction among NPs and more thermally stable than the longer region. The design of these regions including length and sequence determined the effective radii of DNA-linked NPs, which also enable to modulate the competition relationship between entropic and enthalpic contributions in assembly process at different temperature. Additionally, a flexor region between above two regions is added to the linker. When deleting the single base adenine and adding a polyethylene glycol oligomer (PEG) to change the rigidity of flexor region, the latter formed a more ordered crystalline structure, which verifies that the greater flexibility is fit for crystallization. The controllable variations in connection method (as the more complex polyvalent conjugates of DNA and AuNPs, Figure 3C) aim at changing the hydrodynamic radii of DNA-NPs and the coordination environment of NPs, which are presented in one of the six rules for assembly of NPs superlattice [8]. Apart from the above factors, highly monodisperse NPs are also important.
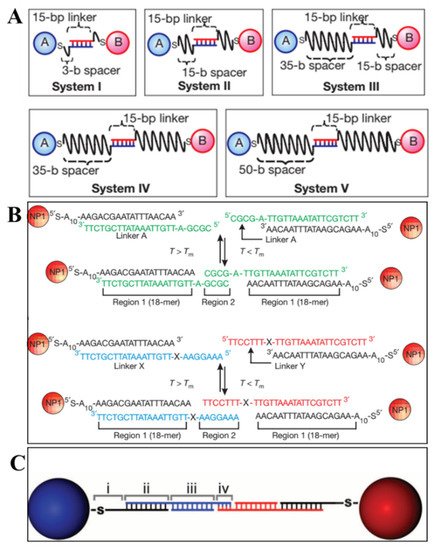
Figure 3. SsDNA linkages between NPs. (A) Connection by two linkers on the surface of NPs (System I to V: different spacers in length) (Adapted with permission from [6]. Copyright 2008, Springer Nature.); (B) connection by a free linker including a flexor region (X region) (Adapted with permission from [7]. Copyright 2008, Springer Nature.); (C) polyvalent conjugates of DNA and AuNPs by four free linkers. (Adapted with permission from [8]. Copyright 2011, American Association for the Advancement of Science).
2.2. Connection via DNA Tile
DNA tile, derived from Holliday junction like the four-arm junction [9] in Figure 4A, consists of several stoichiometric single oligonucleotide strands. In order to assemble larger architecture, a crossover was introduced to design a rigid “DNA double-crossover (DX) molecule” as the building block. On both sides of the DX molecule, a single-strand DNA extended as sticky end for connecting other DX molecules. Based on the different DX motifs and above connection method, some 2D arrays [10][11][12], highly complex 3D nanostructures [13], were constructed and also employed to arrange NPs and proteins.
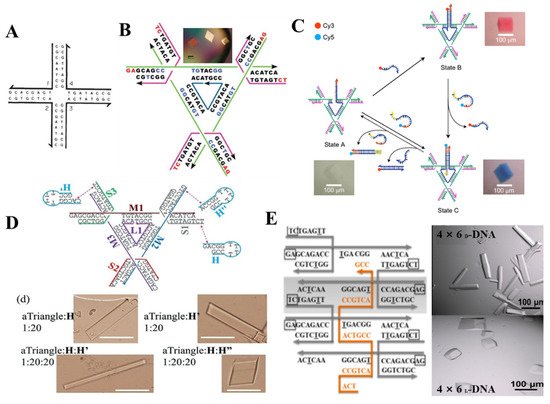
Figure 4. 3D superlattice via DNA tile connection. (A) four-arm junction; (Adapted with permission from [9]. Copyright 1983, Springer Nature). (B) Design of tensegrity triangle as building block and the corresponding assembly, (Adapted with permission from [14]. Copyright 2009, Springer Nature.); (C) Three-state 3D device composed of 3D crystalline lattice and dynamic device (Adapted with permission from [15]. Copyright 2017, Springer Nature.); growth modulation for 3D lattice by agent strand in triangle motif (D) (Adapted with permission from [16]. Copyright 2018, WILEY−VCH Verlag GmbH & Co. KGaA, Weinheim, Germany). A weaving strand in 4 × 5 motif (E) (Adapted with permission from [17]. Copyright 2017, American Chemical Society).
Though the position of NPs could be controlled precisely by DNA array or 3D DNA architecture assembled by DNA tile, there still are some obstacles to form a 3D crystal on the linear, planar template or even polyhedral frame. A groundbreaking work was published by Seeman that a single crystal was obtained by designing a tensegrity triangle [14]. This rigid triangle motif is composed of seven strands (three magenta strands form crossovers in the corners, three green strands extend for the length of each helix, one blue nicked strand at the center), in which the sequences of three magenta and green strands are same respectively, and the blue strand contains triply repeating sequence, a red short single-stranded sticky end was attached at the end of the six helices to connect with six other motifs, thereby yielding rhombohedral 3D crystal (Figure 4B). Afterward, they reported a three-state 3D device by combining devices and 3D crystalline lattice in one system [15] (Figure 4C). By designing the binding of coloring strand and central bule strand, the transition between blue and red crystal could be realized. Moreover, more efforts were made to modulate the growth, the cavity size and chirality, stability of 3D crystal by introducing an agent strand [16] (Figure 4D) at the end of the triangle motif, a weaving strand [17] in the center of a 4 × 5 motif (Figure 4E), enzymatic ligation [18] at position of the sticky end. To design 3D DNA crystals with higher resolution, a series of factors including sticky end length and sequences and 5′/3′ phosphates were systemically analyzed [19]. These advances make it feasible to extend 2D DNA array to 3D crystal, but the dimension of 3D crystal is still limited within a certain range, therefore more effective strategies are desired to break this obstacle.
2.3. Connection via DNA Origami Nanostructure
DNA origami was first termed by Rothemund in 2006 [20], in which a long scaffold DNA with more than 7000 bases and hundreds of short staple strands assembled into anticipated structures ranging from simple 2D planar structure [20] to intricate 3D architectures [21][22][23] (Figure 5A). Over the past four decades, numerous design rules and assembly strategies have been developed to increase the complexity, advancing the structural DNA nanotechnology. The potential applications of these complex system turn to be of importance and attracts a lot of attention. For instance, DNA origami-based nanostructures were employed in biosensor [24][25][26][27], biocomputing [28][29][30][31], biomedicine [32] fields. Recently, DNA origami-guided 3D lattice engineering of inorganic NPs became one of the research hotspots.
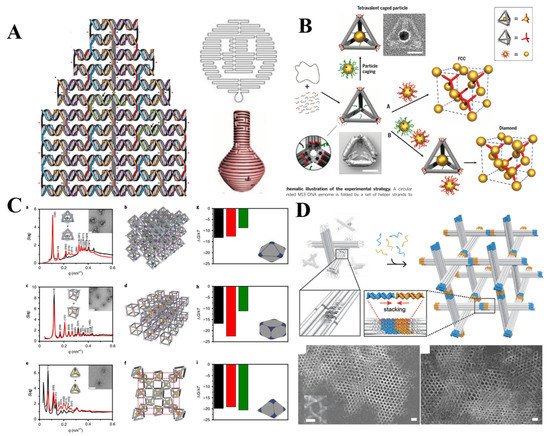
Figure 5. (A) Design of DNA origami and DNA origami-based simple 2D planar structure and intricate 3D architectures. (Adapted with permission from [20]. Copyright 2006, Springer Nature.) (Adapted with permission from [22]. Copyright 2011, American Association for the Advancement of Science); (B) face-centered-cubic and diamond lattice guided by DNA origami tetrahedron. (Adapted with permission from [33]. Copyright 2016, American Association for the Advancement of Science); (C) DNA origami tetrahedron, octahedron, cubic with different valances were employed to engineer diamond type, simple cubic and body-centered-cubic 3D AuNPs lattices. (Adapted with permission from [34]. Copyright 2020, Springer Nature.); (D) DNA origami-based structure assembled into 3D rhombohedral crystal. (Adapted with permission from [35]. Copyright 2018, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).
Initially, NPs could be arranged to prescribed nanoclusters and low-dimensional arrays via the connection of the DNA frame origami structure [36]. A DNA origami octahedron frame is designed based on DNA origami technology, its six vertices can be encoded by extending the different sticky ends, thus determining the NP placement. The successful connection of DNA frame origami structure for 1D and 2D arrays meant it is possible to extend low-dimensional arrays to 3D lattice. One year later, Tian et al. realized above envision through designing the geometrical shapes and matching the topological connection among NPs by frame vertices. A series of DNA polyhedral frames (like cube, octahedron, elongated square bipyramid, prism, and triangular bipyramid) as topological linkers guided the assembly of 3D lattice with different lattice types and parameters such as simple cubic, face-centered-cubic, body-centered-tetragonal, simple hexagonal [37]. This approach sheds new light upon 3D lattice engineering. Liu et al. created a AuNPs diamond superlattice by tetrahedra with low packing fraction, orientation sensitivity and heterogeneity [33] (Figure 5B). The difficulty in diamond lattice assembly was successfully overcome by turning into the assembly between isotropic NP and anisotropic NPs with tetravalent binding topology.
3. Future direction
A full understanding of the 3D superlattice assembly by further investigating the assembly kinetics and arbitrarily controlling the crystal parameters like symmetry, the size, and shape of crystal habit are still required. Combining the fresh ideas of bottom-up DNA-grated 3D superlattice assembly with traditional postprocessing will be a promising direction of 3D lattice-based nanofabrication and further promote the application in real world.
References
- Noguchi, T.; Roy, B.; Yoshihara, D.; Sakamoto, J.; Yamamoto, T.; Shinkai, S. A Chiral Recognition System Orchestrated by Self-Assembly: Molecular Chirality, Self-Assembly Morphology, and Fluorescence Response. Angew. Chem. Int. Ed. 2017, 56, 12518–12522.
- Damasceno, P.F.; Engel, M.; Glotzer, S.C. Predictive Self-Assembly of Polyhedra into Complex Structures. Science 2012, 337, 453–457.
- Pinheiro, A.V.; Han, D.; Shih, W.M.; Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772.
- Bates, F.S.; Hillmyer, M.A.; Lodge, T.P.; Bates, C.M.; Delaney, K.T.; Fredrickson, G.H. Multiblock Polymers: Panacea or Pandora’s Box? Science 2012, 336, 434–440.
- Cheng, H.F.; Wang, S.; Mirkin, C.A. Electron-Equivalent Valency through Molecularly Well-Defined Multivalent DNA. J. Am. Chem. Soc. 2021, 143, 1752–1757.
- Nykypanchuk, D.; Maye, M.M.; van der Lelie, D.; Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 2008, 451, 549–552.
- Park, S.Y.; Lytton-Jean, A.K.R.; Lee, B.; Weigand, S.; Schatz, G.C.; Mirkin, C.A. DNA-programmable nanoparticle crystallization. Nature 2008, 451, 553–556.
- Macfarlane, R.J.; Lee, B.; Jones, M.R.; Harris, N.; Schatz, G.C.; Mirkin, C.A. Nanoparticle Superlattice Engineering with DNA. Science 2011, 334, 204–208.
- Kallenbach, N.R.; Ma, R.-I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 1983, 305, 829–831.
- He, Y.; Chen, Y.; Liu, H.; Ribbe, A.E.; Mao, C. Self-Assembly of Hexagonal DNA Two-Dimensional (2D) Arrays. J. Am. Chem. Soc. 2005, 127, 12202–12203.
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires. Science 2003, 301, 1882–1884.
- He, Y.; Tian, Y.; Ribbe, A.E.; Mao, C. Highly Connected Two-Dimensional Crystals of DNA Six-Point-Stars. J. Am. Chem. Soc. 2006, 128, 15978–15979.
- He, Y.; Ye, T.; Su, M.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201.
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77.
- Hao, Y.; Kristiansen, M.; Sha, R.; Birktoft, J.J.; Hernandez, C.; Mao, C.; Seeman, N.C. A device that operates within a self-assembled 3D DNA crystal. Nat. Chem. 2017, 9, 824–827.
- Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. Modulating Self-Assembly of DNA Crystals with Rationally Designed Agents. Angew. Chem. Int. Ed. 2018, 57, 16529–16532.
- Simmons, C.R.; Zhang, F.; MacCulloch, T.; Fahmi, N.; Stephanopoulos, N.; Liu, Y.; Seeman, N.C.; Yan, H. Tuning the Cavity Size and Chirality of Self-Assembling 3D DNA Crystals. J. Am. Chem. Soc. 2017, 139, 11254–11260.
- Li, Z.; Liu, L.; Zheng, M.; Zhao, J.; Seeman, N.C.; Mao, C. Making Engineered 3D DNA Crystals Robust. J. Am. Chem. Soc. 2019, 141, 15850–15855.
- Ohayon, Y.P.; Hernandez, C.; Chandrasekaran, A.R.; Wang, X.; Abdallah, H.O.; Jong, M.A.; Mohsen, M.G.; Sha, R.; Birktoft, J.J.; Lukeman, P.S.; et al. Designing Higher Resolution Self-Assembled 3D DNA Crystals via Strand Terminus Modifications. ACS Nano 2019, 13, 7957–7965.
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302.
- Ke, Y.G.; Douglas, S.M.; Liu, M.H.; Sharma, T.; Cheng, A.C.; Leung, A.; Liu, Y.; Shih, W.M.; Yan, H. Multilayer DNA Origami Packed on a Square Lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908.
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science 2011, 332, 342–346.
- Han, D.; Pal, S.; Yang, Y.; Jiang, S.; Nangreave, J.; Liu, Y.; Yan, H. DNA Gridiron Nanostructures Based on Four-Arm Junctions. Science 2013, 339, 1412–1415.
- Wang, S.; Zhou, Z.; Ma, N.; Yang, S.; Li, K.; Teng, C.; Ke, Y.; Tian, Y. DNA Origami-Enabled Biosensors. Sensors 2020, 20, 6899.
- Wang, S.; Lu, S.; Zhao, J.; Huang, J.; Yang, X. Real-Time Study of the Interaction between G-Rich DNA Oligonucleotides and Lead Ion on DNA Tetrahedron-Functionalized Sensing Platform by Dual Polarization Interferometry. Acs Appl. Mater. Interfaces 2017, 9, 41568–41576.
- Wang, S.; Lu, S.; Zhao, J.; Ye, J.; Huang, J.; Yang, X. An electric potential modulated cascade of catalyzed hairpin assembly and rolling chain amplification for microRNA detection. Biosens. Bioelectron. 2019, 126, 565–571.
- Wang, S.; Zhao, J.; Lu, S.; Sun, J.; Yang, X. A duplex connection can further illuminate G-quadruplex/crystal violet complex. Chem. Commun. 2019, 55, 1911–1914.
- Petersen, P.; Tikhomirov, G.; Qian, L. Information-based autonomous reconfiguration in systems of interacting DNA nanostructures. Nat. Commun. 2018, 9, 1–10.
- Wang, S.; Sun, J.; Zhao, J.; Lu, S.; Yang, X. Photo-Induced Electron Transfer-Based Versatile Platform with G-Quadruplex/Hemin Complex as Quencher for Construction of DNA Logic Circuits. Anal. Chem. 2018, 90, 3437–3442.
- Wang, S.; Lu, S.; Zhao, J.; Yang, X. A Ratiometric Fluorescent DNA Radar Based on Contrary Response of DNA/Silver Nanoclusters and G-Quadruplex/Crystal Violet. ACS Appl. Mater. Interfaces 2019, 11, 25066–25073.
- Wang, S.; Zhao, J.; Lu, S.; Huang, J.; Yang, X. Establishment of logic gates based on conformational changes in multiple-factor biomolecule interaction process by dual polarization interferometry. Anal. Chem. 2019, 91, 6971–6975.
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.W.; Lin, L.; Qiu, D.L.; Wang, Z.G.; Zou, G.Z.; Liang, X.J.; Yan, H.; et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403.
- Liu, W.; Tagawa, M.; Xin, H.L.; Wang, T.; Emamy, H.; Li, H.; Yager, K.G.; Starr, F.W.; Tkachenko, A.V.; Gang, O. Diamond family of nanoparticle superlattices. Science 2016, 351, 582–586.
- Tian, Y.; Lhermitte, J.R.; Bai, L.; Vo, T.; Xin, H.L.; Li, H.; Li, R.; Fukuto, M.; Yager, K.G.; Kahn, J.S.; et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 2020, 19, 789–796.
- Zhang, T.; Hartl, C.; Frank, K.; Heuer-Jungemann, A.; Fischer, S.; Nickels, P.C.; Nickel, B.; Liedl, T. 3D DNA Origami Crystals. Adv. Mater. 2018, 30.
- Tian, Y.; Wang, T.; Liu, W.; Xin, H.L.; Li, H.; Ke, Y.; Shih, W.M.; Gang, O. Prescribed Nanoparticle Cluster Architectures and LowDimensional Arrays. Nat. Nanotechnol. 2015, 10, 637–644.
- Tian, Y.; Zhang, Y.; Wang, T.; Xin, H.L.; Li, H.; Gang, O. Lattice engineering through nanoparticle–DNA frameworks. Nat. Mater. 2016, 15, 654–661.
More
Information
Subjects:
Genetics & Heredity
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revision:
1 time
(View History)
Update Date:
28 Jul 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




