
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lu Zhang | + 2751 word(s) | 2751 | 2021-06-22 12:10:47 | | | |
| 2 | Lindsay Dong | Meta information modification | 2751 | 2021-07-23 09:49:30 | | |
Video Upload Options
RNA helicases constitute a large family of proteins with functions in all aspects of RNA metabolism, including unwinding or annealing of RNA molecules to regulate pre-mRNA, rRNA and miRNA processing, clamping protein complexes on RNA, or remodeling ribonucleoprotein complexes, to regulate gene expression. RNA helicases also regulate the activity of specific proteins through direct interaction. Abnormal expression of RNA helicases has been associated with different diseases, including cancer, neurological disorders, aging, and autosomal dominant polycystic kidney disease (ADPKD) via regulation of a diverse range of cellular processes such as cell proliferation, cell cycle arrest, and apoptosis.
1. Introduction
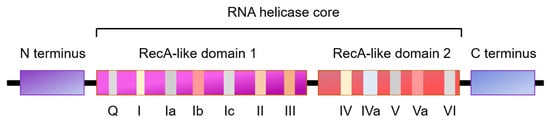
2. The Role of RNA Helicases in Regulation of Cell Cycle Progression
2.1. RNA Helicases Regulate G1-S Phase Transition
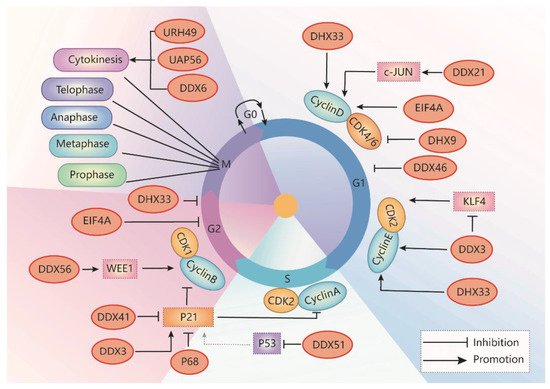
DDX3/Ded1 is one of the most widely studied DEAD-box RNA helicases. DDX3 regulates RNA metabolism, including transcription, pre-mRNA splicing, RNA export, and translation, which plays a critical role in many biological processes [9]. DDX3 also promotes phase separation when it interacts with ATP [10]. DDX3 also regulates cell apoptosis via the p53 signaling pathway during embryonic development in animal models [11]. Dysregulation of DDX3 plays a critical role in various diseases, including inflammation, viral infection, neurological disorders, and cancer [10][12]. The role of DDX3 in oncogenesis is related to the regulation of cell growth, cell cycle, and cell survival. Increased DDX3 expression promotes cancer cell growth in medulloblastoma, colorectal, breast, prostate, and lung cancer [9][13][14][15], whereas depletion of DDX3 induces cell cycle arrest in the G1 phase in cancer cells of those cancers (Table 1) [9][16][17][18][19]. Knockdown of DDX3 also inhibits cell cycle progression by blocking entry into the S phase. Mechanically, DDX3 facilitates translation initiation of the cell cycle regulator cyclin E1 mRNA via its 5′ UTR [20]. In addition, DDX3 inhibits the expression of Krüppel-like factor 4 (Klf4) by altering its mRNA splicing, resulting in an upregulation of the expression of CCNA2 and CDK2 [16]. In sum, DDX3 promotes G1/S transition by promoting cyclin E1 translation, suppressing KLF4 expression, and promoting CDK2 expression (Figure 2).
| Cell Cycle Stage | RNA Helicases | Intracellular Location | Expression |
|---|---|---|---|
| G1-S phase transition | Ded1/DDX3 | Nuclear speckles and cytoplasm | Upregulation in medulloblastoma, colorectal, breast, prostate, and lung cancer |
| DHX33 | Nucleus and nucleoli | Upregulation in lung cancers, hepatocellular carcinoma, lymphoma, colon cancer, and glioblastoma | |
| DHX9 | Nucleus | Upregulation in cervical cancer, breast cancer, prostate cancer, colorectal cancer, hepatocellular carcinoma, and Ewing sarcoma | |
| DDX21 | Nucleus and cytoplasm | Dysregulation in colon cancer, lymphomas, neuroblastoma, and breast cancers | |
| eIF4A | Nucleus and cytoplasm | Dysregulation in pancreatic cancer, breast cancer, prostate cancer | |
| DDX46 | Focal nuclear | Upregulated in colorectal carcinoma, esophageal squamous cell carcinoma, gastric cancer, and osteosarcoma cells | |
| S phase progression | DDX51 | Predominantly in nuclear | Dysregulation in NSCLC |
| G2-M phase transition | DDX56 | Nucleolus | Upregulation of DDX56 in various cancer, including osteosarcoma, colorectal cancer, and relates to a poor prognosis |
| DHX33 | Nucleus and nucleoli | See above | |
| Mitosis | UAP56 | Nucleus and cytoplasm | Not clear |
| URH49 | Nucleus and cytoplasm | Not clear | |
| Cytokinesis | UAP56 | Nucleus and cytoplasm | Not clear |
| URH49 | Nucleus and cytoplasm | Not clear | |
| DDX6 | Nucleus and cytoplasm | colorectal cancer and hepatocellular carcinoma | |
| Regulate the expression of p21(WAF1/CIP1) | DDX41 | Nucleus and cytoplasm | DDX41 mutant leads to anemia and acute myeloid leukemia. DDX41 increased in cervical cancer. |
| DDX5 | Mostly in nucleus, cytoplasmic levels of DDX5 increased in the G2/M phase | p68 increased in a range of cancers except for hepatocellular carcinoma | |
| DDX3 | Predominantly in nuclear speckles and at low levels in cytoplasm | See above |
2.2. RNA Helicases Regulate S Phase Progression
During the S phase of the cell cycle, genetic materials, such as DNA, are synthesized for the duplication of the chromatin and reproduction of the whole genome, which is a pre-requisition for cell division. Once cells enter the S phase, cyclin E/CDK2 complex needs to be silenced to eliminate the DNA re-replication [21]. Dissociation of CDK2 from cyclin E/CDK2 complex results in its association with the newly synthesized cyclin A to form CDK2/cyclin A complex, which can phosphorylate proteins that are necessary for completion of the S phase. Alternatively, spliced p53 isoform (Delta-p53) transactivates the expression of endogenous p21, resulting in the inhibition of CDK2/cyclin A activity to and attenuate S phase progression [22].
2.3. RNA Helicases Regulate G2/M Phase Transition
2.4. RNA Helicases Regulate Cytokinesis
Cytokinesis is the last step of the cell cycle in which the cell must devotedly split the chromosomes and cytoplasm to produce two daughter cells with equal contents [27]. RNA helicases are involved in the key steps of cytokinesis, including the assembly and contraction of the contractile network, the formation of the mitotic spindle, and the interaction between the microtubule and cortical actomyosin cytoskeleton [28].
2.5. RNA Helicases Target CDK Inhibitor p21
3. Development of RNA Helicase Inhibitors for Clinical Treatment
3.1. Different Targeting Strategies Are Used to Develop Compounds against RNA Helicases
The important roles of RNA helicases in the regulation of the cell cycle, cell proliferation, cellular transformation, apoptosis, and cell adhesion and motility make RNA helicases an active targets of drug development for the treatment of viral infections, neurodegenerative diseases, and cancers. Based on the crystal structures and the mechanism(s) of activity of the enzymes, specific and selective inhibitors for RNA helicases have been designed. In general, inhibitors targeting RNA helicase activity could act via one or more of the mechanisms: (1) inhibition of the ATPase activity of RNA helicases by interference with ATP binding and result in limiting the energy necessary for the unwinding and translocation; (2) inhibition of the helicase activity by competitively occupying the RNA binding site of the RNA helicases; and (3) stabilization of RNA helicases onto RNA, resulting in the inhibition of translation initiation (Table 2).
| RNA Helicases | Inhibitors | Chemical Structure | Mechanisms of Action | Diseases | Model | Toxicity or Tissue-Specific |
|---|---|---|---|---|---|---|
| DDX3 | RK-33 | 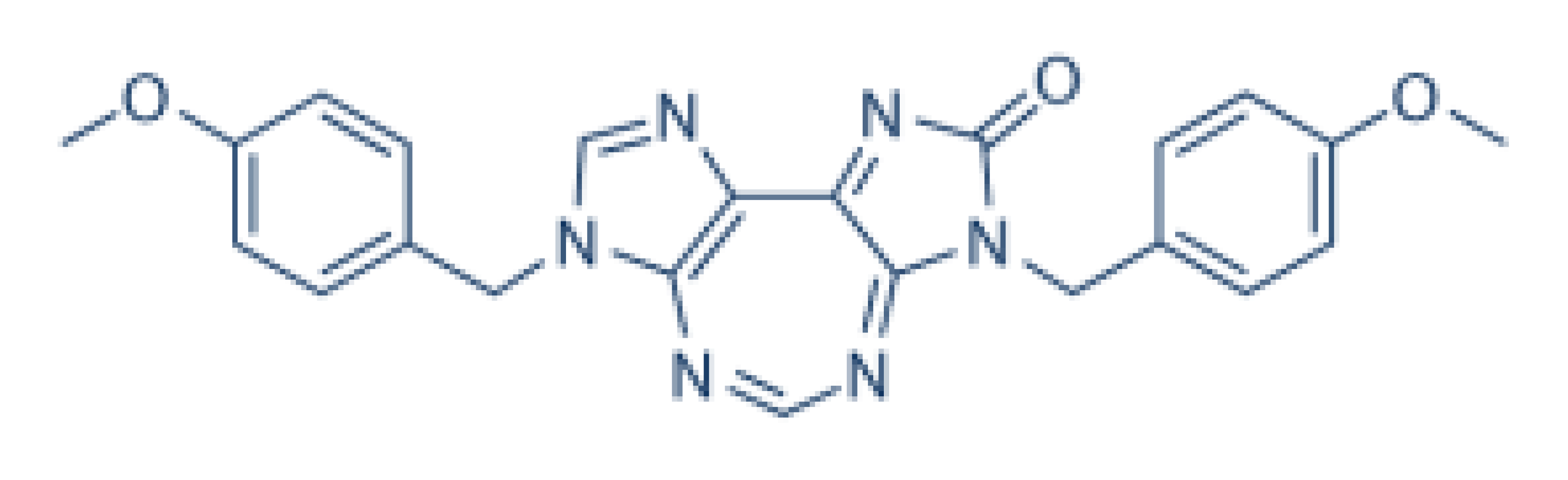 |
Inhibition of helicase activity | Lung cancer, medulloblastoma, prostate cancer, Ewing sarcoma, and colorectal cancer | In vitro, and animal models | No discernable toxicity in animal models |
| NZ51 | 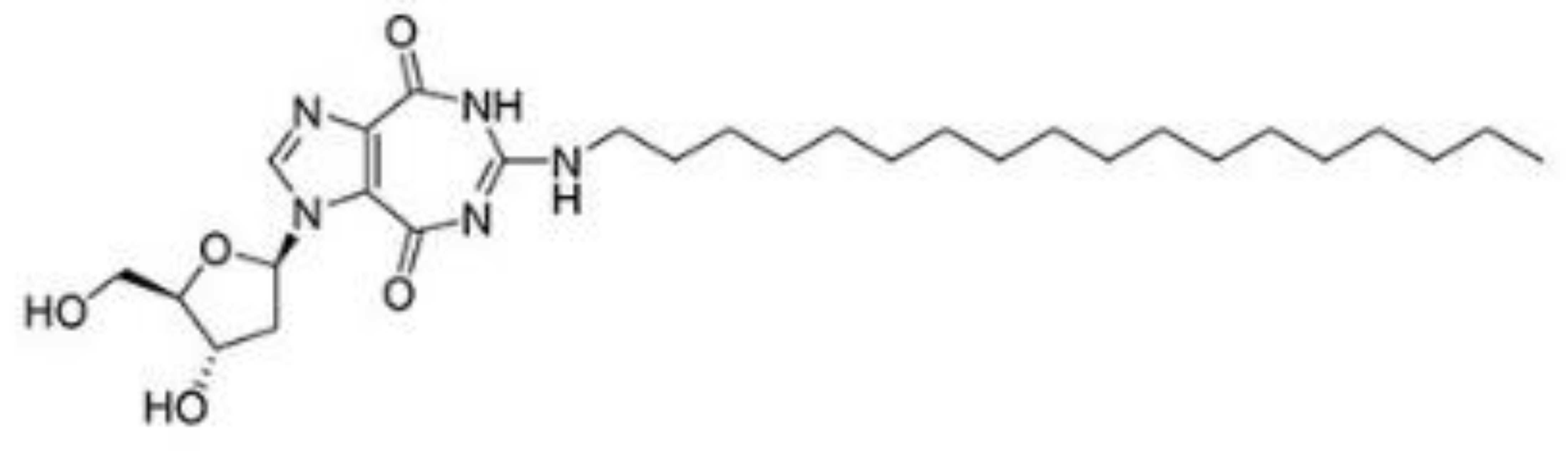 |
Inhibition of helicase activity | Breast cancer | In vitro | Not clear | |
| Doxorubicin | 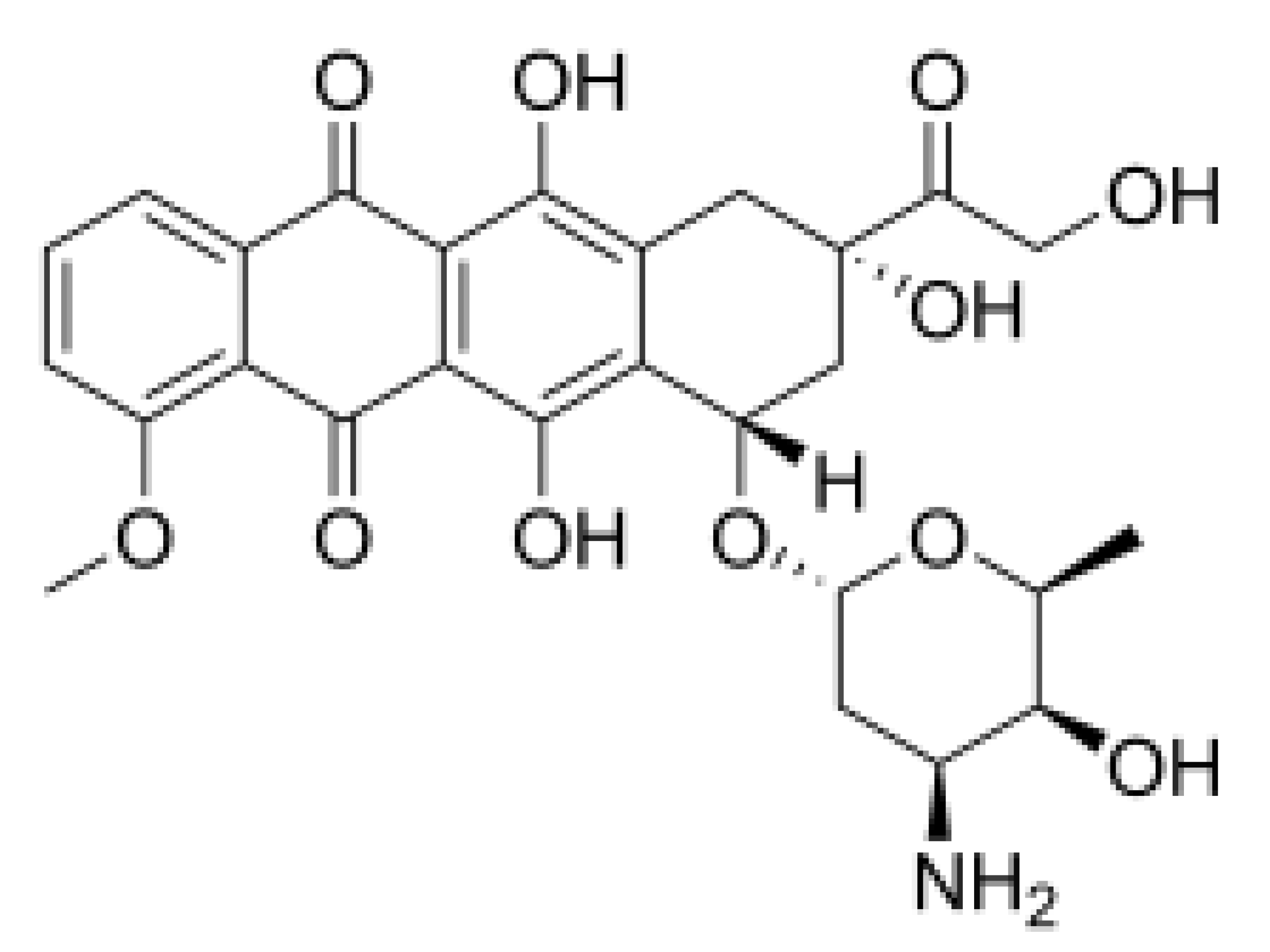 |
Inhibition of ATPase activity | Oral squamous cell carcinoma | In vitro | Cardiotoxicity | |
| eIF4A | Compound 18 | 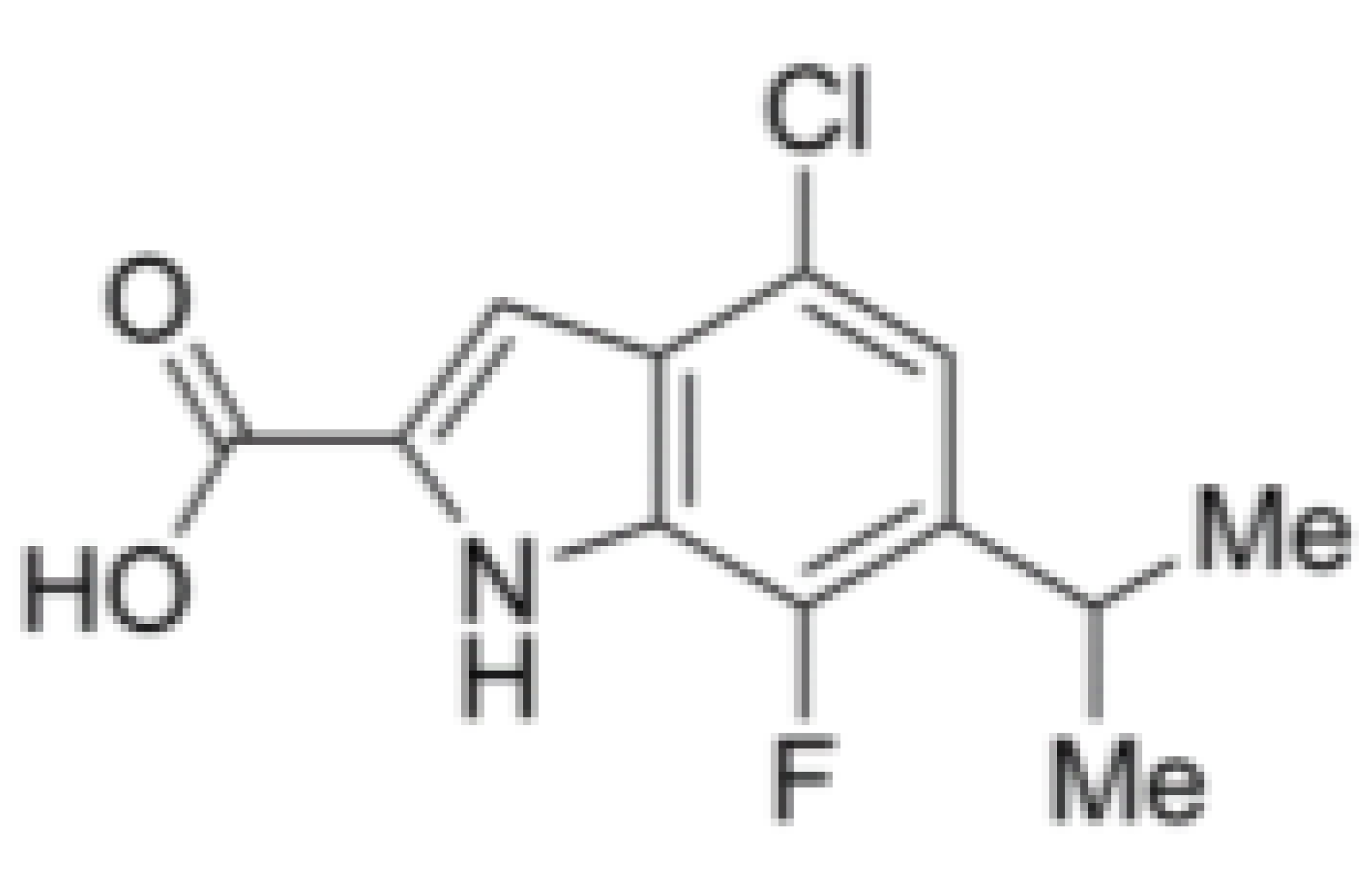 |
Inhibition of ATPase activity | Exon junction complex | NA | IC50: 0.97 μmol/L |
| Silvestrol | 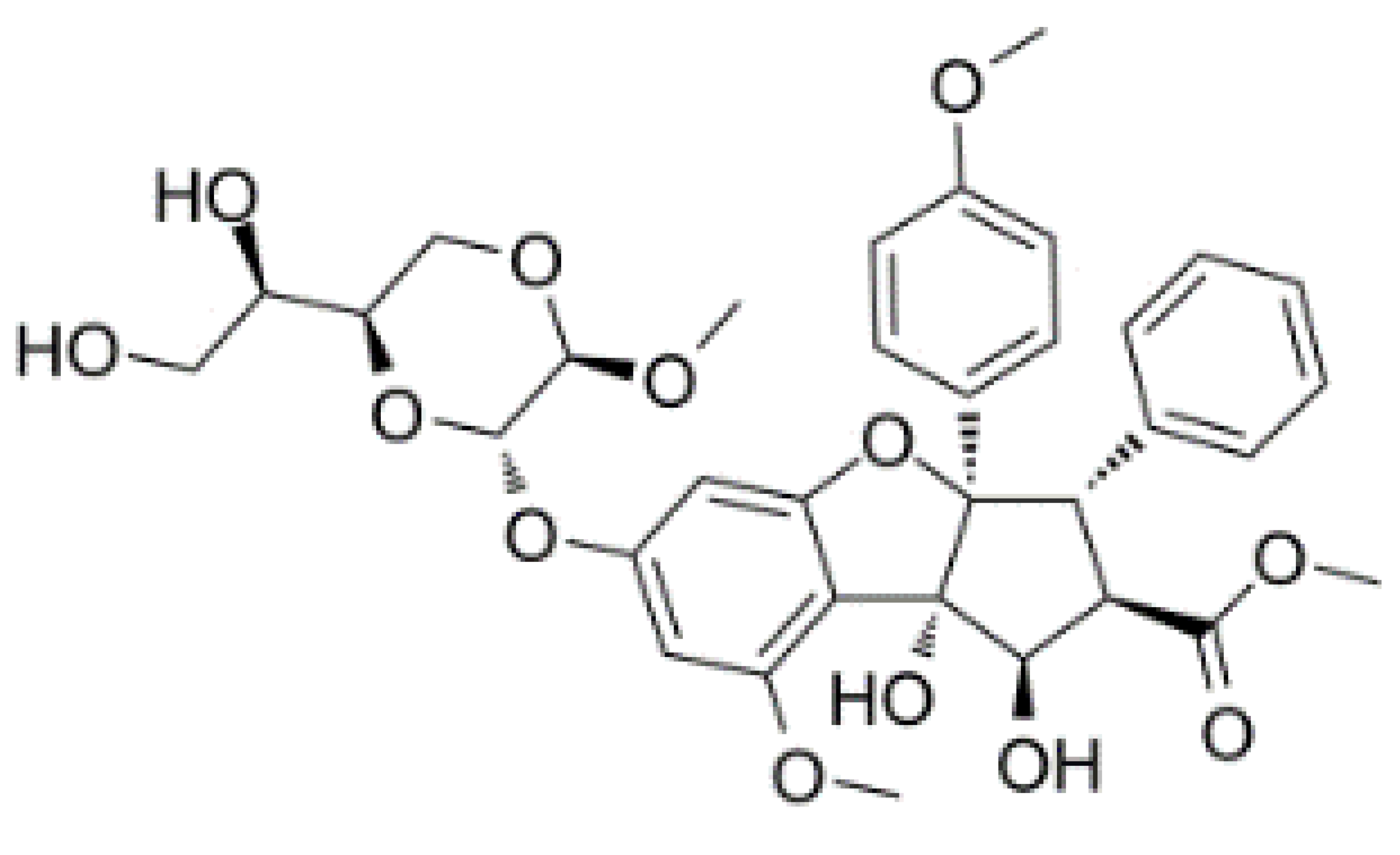 |
Stabilization of RNA helicase onto RNA | Breast cancer | In vitro | Not toxic in vitro and in vivo at concentrations of effective activity | |
| Hippuristanol | 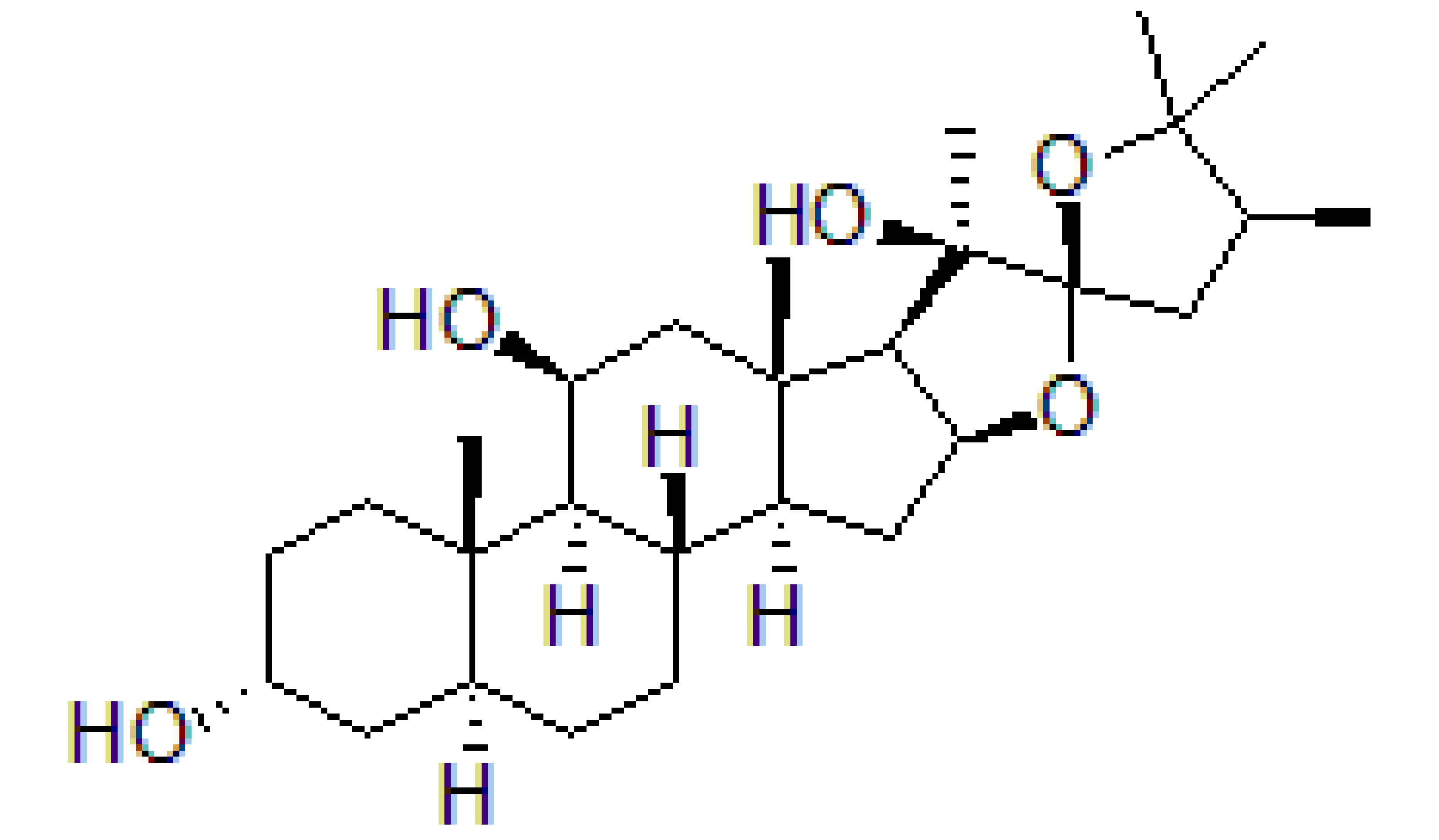 |
Inhibition of helicase activity | Leukemia | In vitro | Not clear | |
| CR-1-31-B | 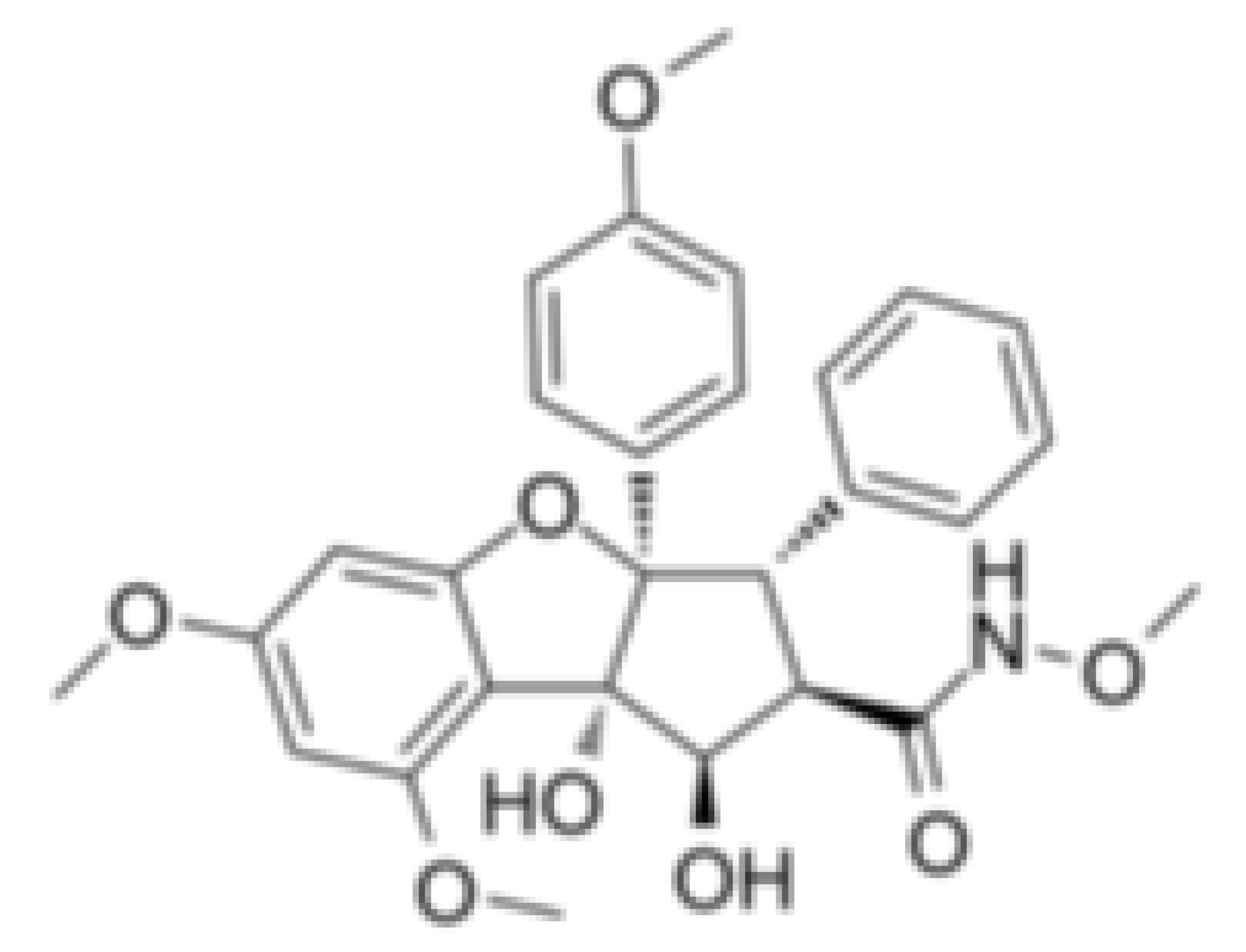 |
Stabilization of RNA helicase onto RNA | Breast cancer | In vitro | Not clear | |
| Pateamine A | 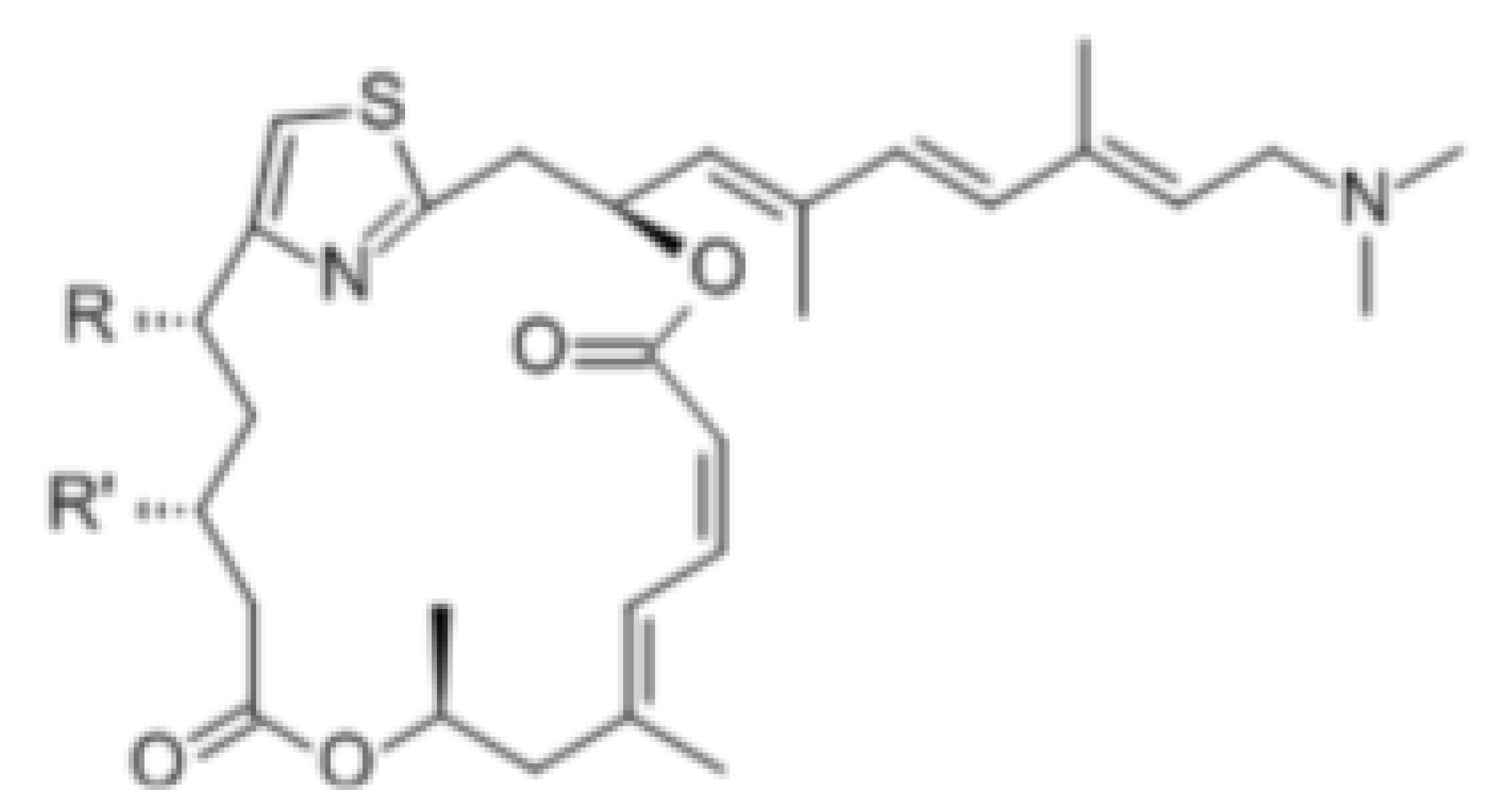 |
Regulation of ATPase and RNA helicase activity | Melanoma | In vitro, and animal models | Low toxicity to quiescent cells | |
| DDX6 | RX-5902 | 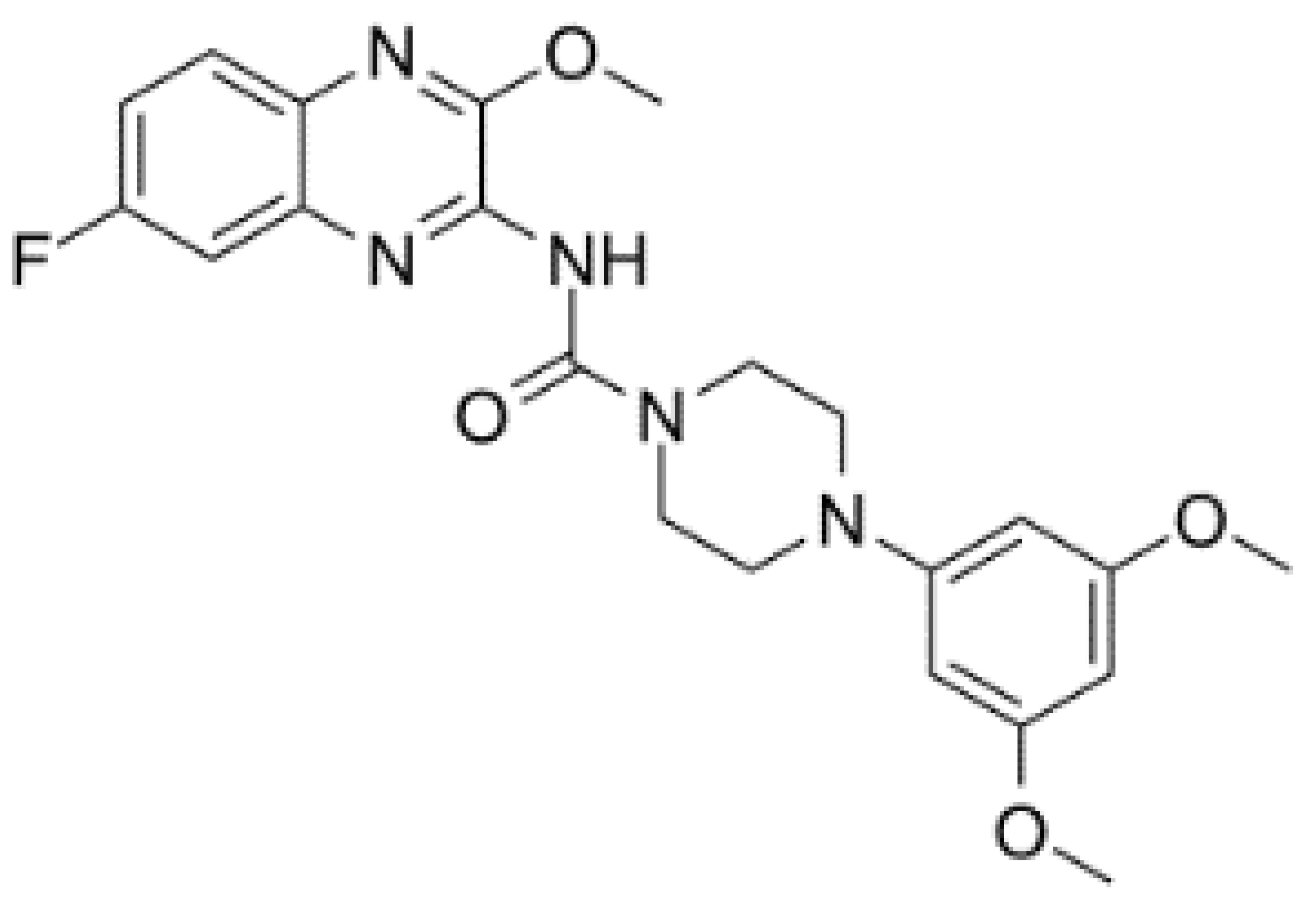 |
Inhibition of ATPase activity | TNBC | In vitro, preclinical models of TNBC, phase I study | Not clear |
3.2. The Therapeutic Potential of RNA Helicase Inhibitors in the Treatment of Various Disorders
As described above, different inhibitors of RNA helicases have been developed, which may potentially be used in disease treatment [17][34][35]. The DDX3 inhibitor, NZ51, is able to block the ATP-dependent helicase activity of DDX3. NZ51 suppresses DDX3 activity at low micromole concentration and displays anti-proliferative activity by inhibiting cell replication at the G1 phase in aggressive breast cancer cells [12][19]. However, treatment with NZ51 did not significantly reduce tumor growth in breast cancer mouse model [19]. Doxorubicin, an antitumor drug, can inhibit the ATPase activity of DDX3, which shows anticancer activity on human oral squamous cell carcinoma cells. However, the cardiotoxicity is a limitation for doxorubicin in vivo [36]. The DDX3 inhibitors, RK-33, can bind with the ATP-binding domain of DDX3 to block its helicase activity.
Hippuristanol, as a pan eIF4A inhibitor, has been reported to inhibit human T lymphotrophic virus type 1-infected T-cell line and adult T-cell leukemia cell proliferation by promoting cell cycle arrest at the G1 phase and suppressing the expression of cell cycle proteins and cyclin-dependent kinases, and inducing apoptosis by decreasing the expression levels of Bcl-x, baculoviral IAP repeat, containing 3 X-linked inhibitors of apoptosis (xIAP) and caspase 8 and FADD like apoptosis regulator [37][38].
DDX5 (p68) can be phosphorylated at the Y593 site by growth factors, including platelet-derived growth factor, to increase cellular proliferation, epithelial to mesenchymal transition (EMT), malignant transformation, cell migration, and oncogenesis through translocation of β-catenin to nuclear and activation of cyclin D1, c-JUN, and c-MYC [39].
4. RNA Helicases and Phase-Separated Organelles
Recently, several studies have demonstrated that the DEAD-box RNA helicases involved in regulation of liquid–liquid phase separation (LLPS) play a crucial role in the formation of RNA-containing membrane-less organelles, including pericentriolar material (PCM), stress granules (SGs), P bodies, and P granules in Caenorhabditis elegans [40].
Pericentriolar material is defined as a matrix of proteins surrounding the two barrel-shaped centrioles during mitosis, which serves as a platform for protein complexes to regulate organelle trafficking, protein degradation, and spindle assembly.
Stress granules are RNPs that assemble in response to environmental stresses such as oxidative stress, heat shock, or osmotic shock. One component of yeast stress granules is Ded1p, also named DDX3 in mammalian. Temperature-driven liquid phase condensation of Ded1p induces the sequestration of housekeeping mRNAs and promotes an expanded heat shock response in program results of the preferential expression of stress proteins at 39 °C [41].
P bodies are the large cytoplasmic granules, which are membrane-less cytoplasmic organelles that form via phase-separation once RNAs and nearby RBPs assemble into ribonuclear particle (RNP) granules. P bodies have all the key characters of LLPS, which are liquid-like, spherical, and dynamic, and can be dissolved by the alcohol 1,6-hexanediol.
P granules are RNA/protein condensates in the germline of Caenorhabditis elegans. P granules are membrane-less organelles that may assemble by intracellular phase separation. GLH-1, a germline putative RNA helicase, regulates the formation and disassembly of P granules coupling with distinct steps of its ATPase hydrolysis cycle [42].
DDXs globally promote phase separation in their ATP- and RNA-bound state. ATP hydrolysis induces the release of RNA clients from a DDX, results in the disassembly of RNA-containing membrane-less organelles [40]. The condensates formed by 2NT-DDX4YFP, a recombinant protein, dissolve during mitosis and leads to increasing of noise in protein concentration.
5. Conclusions
References
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37.
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215.
- Jarmoskaite, I.; Russell, R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA 2011, 2, 135–152.
- Cordin, O.; Hahn, D.; Beggs, J.D. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012, 24, 431–438.
- Owttrim, G.W. RNA helicases: Diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013, 10, 96–110.
- Soultanas, P.; Wigley, D.B. Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci. 2001, 26, 47–54.
- Bourgeois, C.F.; Mortreux, F.; Auboeuf, D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 426–438.
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352.
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38.
- Chao, C.H.; Chen, C.M.; Cheng, P.L.; Shih, J.W.; Tsou, A.P.; Lee, Y.H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006, 66, 6579–6588.
- Wu, D.W.; Liu, W.S.; Wang, J.; Chen, C.Y.; Cheng, Y.W.; Lee, H. Reduced p21(WAF1/CIP1) via alteration of p53-DDX3 pathway is associated with poor relapse-free survival in early-stage human papillomavirus-associated lung cancer. Clin. Cancer Res. 2011, 17, 1895–1905.
- Heerma van Voss, M.R.; Kammers, K.; Vesuna, F.; Brilliant, J.; Bergman, Y.; Tantravedi, S. Global Effects of DDX3 Inhibition on Cell Cycle Regulation Identified by a Combined Phosphoproteomics and Single Cell Tracking Approach. Transl. Oncol. 2018, 11, 755–763.
- Tantravedi, S.; Vesuna, F.; Winnard, P.T., Jr.; Martin, A.; Lim, M.; Eberhart, C.G.; Berlinicke, C.; Raabe, E.; van Diest, P.J.; Raman, V. Targeting DDX3 in Medulloblastoma Using the Small Molecule Inhibitor RK-33. Transl. Oncol. 2019, 12, 96–105.
- Fu, W.; Verma, D.; Burton, A.; Swaminathan, S. Cellular RNA Helicase DHX9 Interacts with the Essential Epstein-Barr Virus (EBV) Protein SM and Restricts EBV Lytic Replication. J. Virol. 2019, 93, e01244-18.
- Xie, M.; Vesuna, F.; Tantravedi, S.; Bol, G.M.; Heerma van Voss, M.R.; Nugent, K.; Malek, R.; Gabrielson, K.; van Diest, P.J.; Tran, P.T.; et al. RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3. Cancer Res. 2016, 76, 6340–6350.
- Cannizzaro, E.; Bannister, A.J.; Han, N.; Alendar, A.; Kouzarides, T. DDX3X RNA helicase affects breast cancer cell cycle progression by regulating expression of KLF4. FEBS Lett. 2018, 592, 2308–2322.
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015, 7, 648–669.
- Heerma van Voss, M.R.; Vesuna, F.; Trumpi, K.; Brilliant, J.; Berlinicke, C.; de Leng, W.; Kranenburg, O.; Offerhaus, G.J.; Burger, H.; van der Wall, E.; et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 2015, 6, 28312–28326.
- Xie, M.; Vesuna, F.; Botlagunta, M.; Bol, G.M.; Irving, A.; Bergman, Y.; Hosmane, R.S.; Kato, Y.; Winnard, P.T., Jr.; Raman, V. NZ51, a ring-expanded nucleoside analog, inhibits motility and viability of breast cancer cells by targeting the RNA helicase DDX3. Oncotarget 2015, 6, 29901–29913.
- Lai, M.C.; Chang, W.C.; Shieh, S.Y.; Tarn, W.Y. DDX3 regulates cell growth through translational control of cyclin E1. Mol. Cell Biol. 2010, 30, 5444–5453.
- Malumbres, M.; Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005, 30, 630–641.
- Rohaly, G.; Chemnitz, J.; Dehde, S.; Nunez, A.M.; Heukeshoven, J.; Deppert, W.; Dornreiter, I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell 2005, 122, 21–32.
- Schmidt, M.; Rohe, A.; Platzer, C.; Najjar, A.; Erdmann, F.; Sippl, W. Regulation of G2/M Transition by Inhibition of WEE1 and PKMYT1 Kinases. Molecules 2017, 22, 2045.
- Mueller, P.R.; Leise, W.F., 3rd. Measurement of Wee kinase activity. Methods Mol. Biol. 2005, 296, 299–328.
- Taylor, W.R.; Stark, G.R. Regulation of the G2/M transition by p53. Oncogene 2001, 20, 1803–1815.
- Stewart, Z.A.; Pietenpol, J.A. p53 Signaling and cell cycle checkpoints. Chem. Res. Toxicol. 2001, 14, 243–263.
- Mierzwa, B.; Gerlich, D.W. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell 2014, 31, 525–538.
- Pollard, T.D.; O’Shaughnessy, B. Molecular Mechanism of Cytokinesis. Annu. Rev. Biochem. 2019, 88, 661–689.
- Peters, D.; Radine, C.; Reese, A.; Budach, W.; Sohn, D.; Janicke, R.U. The DEAD-box RNA helicase DDX41 is a novel repressor of p21(WAF1/CIP1) mRNA translation. J. Biol. Chem. 2017, 292, 8331–8341.
- Dai, L.; Liu, Y.; Liu, J.; Wen, X.; Xu, Z.; Wang, Z.; Sun, H.; Tang, S.; Maguire, A.R.; Quan, J.; et al. A novel cyclinE/cyclinA-CDK inhibitor targets p27(Kip1) degradation, cell cycle progression and cell survival: Implications in cancer therapy. Cancer Lett. 2013, 333, 103–112.
- Bali, A.; O’Brien, P.M.; Edwards, L.S.; Sutherland, R.L.; Hacker, N.F.; Henshall, S.M. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin. Cancer Res. 2004, 10, 5168–5177.
- Juan, G.; Ardelt, B.; Li, X.; Mikulski, S.M.; Shogen, K.; Ardelt, W.; Mittelman, A.; Darzynkiewicz, Z. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia 1998, 12, 1241–1248.
- Ando, T.; Kawabe, T.; Ohara, H.; Ducommun, B.; Itoh, M.; Okamoto, T. Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J. Biol. Chem. 2001, 276, 42971–42977.
- Sanghvi, V.R.; Mohan, P.; Singh, K.; Cao, L.; Berishaj, M.; Wolfe, A.L.; Schatz, J.H.; Lailler, N.; de Stanchina, E.; Viale, A.; et al. NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy. Cancers 2021, 13, 639.
- Capasso, A.; Bagby, S.M.; Dailey, K.L.; Currimjee, N.; Yacob, B.W.; Ionkina, A.; Frank, J.G.; Kim, D.J.; George, C.; Lee, Y.B.; et al. First-in-Class Phosphorylated-p68 Inhibitor RX-5902 Inhibits beta-Catenin Signaling and Demonstrates Antitumor Activity in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2019, 18, 1916–1925.
- Botlagunta, M.; Kollapalli, B.; Kakarla, L.; Gajarla, S.P.; Gade, S.P.; Dadi, C.L.; Penumadu, A.; Javeed, S. In vitro anti-cancer activity of doxorubicin against human RNA helicase, DDX3. Bioinformation 2016, 12, 347–353.
- Tsumuraya, T.; Ishikawa, C.; Machijima, Y.; Nakachi, S.; Senba, M.; Tanaka, J.; Mori, N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem. Pharmacol. 2011, 81, 713–722.
- Tsai, B.P.; Jimenez, J.; Lim, S.; Fitzgerald, K.D.; Zhang, M.; Chuah, C.T.; Axelrod, H.; Wilson, L.; Ong, S.T.; Semler, B.L.; et al. A novel Bcr-Abl-mTOR-eIF4A axis regulates IRES-mediated translation of LEF-1. Open Biol. 2014, 4, 140180.
- Yang, L.; Lin, C.; Liu, Z.R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 2006, 127, 139–155.
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148.
- Iserman, C.; Desroches Altamirano, C.; Jegers, C.; Friedrich, U.; Zarin, T.; Fritsch, A.W.; Mittasch, M.; Domingues, A.; Hersemann, L.; Jahnel, M.; et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 2020, 181, 818–831.e19.
- Chen, W.; Hu, Y.; Lang, C.F.; Brown, J.S.; Schwabach, S.; Song, X.; Zhang, Y.; Munro, E.; Bennett, K.; Zhang, D.; et al. The Dynamics of P Granule Liquid Droplets Are Regulated by the Caenorhabditis elegans Germline RNA Helicase GLH-1 via Its ATP Hydrolysis Cycle. Genetics 2020, 215, 421–434.




