
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yukihiro Yoshimura | + 2204 word(s) | 2204 | 2020-05-14 04:26:48 | | | |
| 2 | Nicole Yin | -27 word(s) | 2177 | 2020-10-30 04:33:52 | | |
Video Upload Options
Mass spectrometry imaging (MSI) or imaging mass spectrometry (IMS) is a promising analytical method to determine and visualize the spatial distribution of specific chemical compositions by their molecular weights or m/z values. MSI is basically a two-dimensional analysis method that can detect intact molecules within tissues or tissue sections without necessitating extraction, purification, and separation.
1. Introduction
Consuming food is essential for survival, maintaining health, and triggering positive emotions like pleasure. One of the factors that drive us toward such behavior is the presence of various compounds in foods. There are many methods to analyze these molecules in foods; however, it is difficult to analyze the spatial distribution of these compounds using conventional techniques, such as mass spectrometry combined with high-performance liquid chromatography or gas chromatography. Mass spectrometry imaging (MSI) is a two-dimensional ionization technology that enables detection of compounds in tissue sections without extraction, purification, separation, or labeling. There are many methods for ionization of analytes, including secondary ion mass spectrometry, matrix-assisted laser desorption/ionization, and desorption electrospray ionization. Such MSI technologies can provide spatial information on the location of a specific analyte in food. The number of studies utilizing MSI technologies in food science has been increasing in the past decade.
The overall methodology behind MSI is quite simple (Figure 1A). In MSI, different layers present on sample’s surface is subsequently analyzed by scraping it with a laser beam irradiation from instrument platform. The spatial resolution is controlled by the operator and is limited by the laser spot size which is different for different instrument platforms, and many spectra are collected at specific coordinates of a sample surface. Then, all spectra are compiled into one data, wherein m/z values of interest can be selected, and their spatial distribution depicted as an image. Different ionization methods have been used in food science, including secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption/ionization (MALDI), and desorption electrospray ionization (DESI), as well as other methods (Figure 1B).
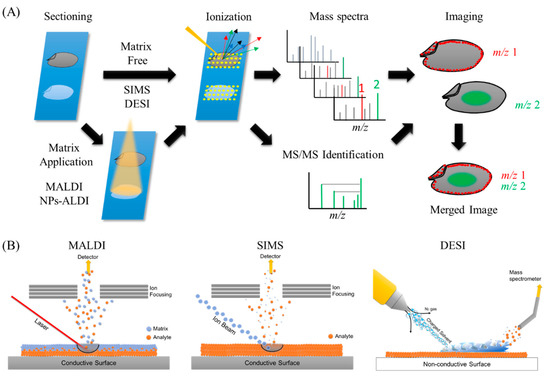
Figure 1. Ionization processes of the most widely used methods in mass spectrometry imaging (MSI) and the general overview of MSI workflow. (A) General overview of MSI workflow. Thin-section samples are mounted on a plate, coated with or without matrix. Then, by using a laser (matrix-assisted laser desorption/ionization (MALDI)), ion beam (secondary ion mass spectrometry (SIMS)), or charged solvent droplets (desorption electrospray ionization (DESI)), ions are produced which are detected by a mass spectrometer. Mass spectra are obtained from every single data point. These data are compiled into a single dataset where the occurrence of any single mass can be visualized as a scaled false color. Molecules of interest can be visualized and identified by MS/MS on tissues. (B) Ionization mechanisms involved in MALDI, SIMS, and DESI ionization sources.
This entry provides an overview of some of the recent applications of MSI in food science and related fields. In the future, MSI will become one of the most promising technologies for visualizing the distribution of food components and for identifying food-related factors by their molecular weights to improve quality, quality assurance, food safety, nutritional analysis, and to locate administered food factors.
2. History of MSI and Widely Used Ionization Methods
To date, many instrument platforms and ionization methods have been applied to MSI. With regards to the various ionization methods utilized, the three techniques which have been widely used in MSI include SIMS [[1][2][3][4]], MALDI [[5][6]], and DESI [[7][8]]. For the past 60 years, since the advent of MSI, it has been widely applied in biomedical sciences, life sciences, food sciences, nutritional sciences, and also in criminal investigations (analysis of fingerprints) [[9]]. The numbers of published articles containing the words ‘mass’, ‘spectrometry’, and ‘imaging’ in the title of the manuscript or in the abstract in the PubMed database has gradually increased over the past two decades (Figure 2). Among these articles, those that contain the additional word ‘food’ is less than 10%, but these publications have been increasing rapidly over the past decade (Figure 2).
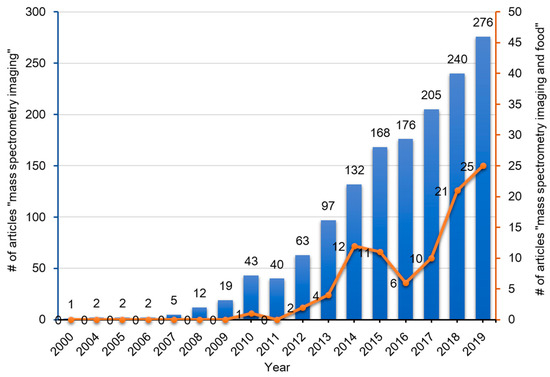
Figure 2. Publication trend (number of published articles) related to mass spectrometry imaging (blue bars) and mass spectrometry imaging of foods (red line). The number of publications were determined by using PubMed for research articles published from 2000–2020 containing the words ‘mass’, ‘spectrometry’, and ‘imaging’ with or without ‘food’ in the title or abstract, respectively.
At the start of MSI history in 1962, SIMS revealed the first ion images, which showed the distribution of Mg+, Al+, and Si+ ions in the specimen [[10]]. In SIMS-MSI experiments, the sample is attached to a conductive surface and subjected to etching by a beam of high-energy primary ions. A wide range of ion sources such as Ar+, Au3+, or Bi3+ can be used in SIMS experiments. Upon collision with the sample surface, the energy of the primary ions is transferred to the analyte to provide secondary ionization (Figure 1B) [[11]]. The primary ion beam allows SIMS instruments to collect nm-sized pixels and to generate high resolution images, but this process is usually limited in the mass ranges of biological samples owing to less molecules being ejected and ionized from the sample surface and also owing to a high degree of ion fragmentation, especially in biological molecules with high molecular weights. Due to these limitations, the use of SIMS in food science is limited to the characterization of small molecule compounds such as elements and lipids, but has the advantage of a very high lateral resolution (>1 µm) compared with other ionization techniques (Table 1).
Table 1. Summary of main ionization methods of MSI.
|
Instrument |
Type of Ionization |
Lateral Resolution |
Applicable |
Favorable Target Molecules |
|
SIMS (secondary ion mass soectrometry) |
Sputtering of sample surface with the impact of a high-energy primary ion |
> 1 µm |
solid |
Elements, fatty acids, lipids, |
|
MALDI (matrix assisted laser desorption/ionization) |
Laser ablation and desorption/ionization within ablated plume |
5-200 µm |
solid |
Metabolites, lipids, peptides, carbohydrates |
|
DESI (desorption electrospray ionization) |
Surface extraction and ionization with highly charged electrospray droplets |
50-200 µm |
solid, liquid, frozen, and gaseous |
Lipids, peptides, small metabolites |
The second ionization technique used for MSI is MALDI, which has been widely used in not only food science [[5][6]], but also in many biological research fields [[12][13][14][15]]. The first report on the utilization of MALDI-MS was by Karas and Hillenkamp in 1988, who performed successful ionization of serum albumin protein [[16]]. In 1997, Caprioli et al. showed the distribution of IB-1 protein fragment (m/z 7605) in an aggregate of human buccal mucosa cells using MALDI-MSI [[17]]. MALDI instruments incorporate a high powered ultraviolet (UV) or infra-red (IR) laser to ionize compounds on the surface of the sample. The matrix, which is typically a low molecular weight compound that easily absorbs laser energy, is coated on the surface of the sample, mixed and crystallized with the molecules of interest, followed by desorption/ionization with a laser beam. Matrix is usually applied to the sample surface by a sprayer or by solvent-free sublimation [[18][19]]. When applying uniform matrix coatings on the surface of the sample, the number of sample types analyzed and their spatial resolution are limited (5–200 µm). However, the utilization of matrix dramatically increases ionization efficiency of compounds, which allows for lower laser intensities resulting in gentle ionization enough to prevent the fragmentation of analytes with high molecular weights. The high ionization efficiency and lower fragmentation of analytes in MALDI-MSI allows for non-targeted and comprehensive analysis of a sample on a slide [[20]]. The analyte, which is easy to ionize, varies according to the type of matrix used (Table 1). In addition, owing to easily absorbing laser energy, the matrix itself sometimes causes ionization interference. Therefore, the choice of the appropriate matrix is very important (Table 2). However, the necessity of selecting matrix suited to the analytes of interest might be time consuming. The detailed methodology of MALDI-MSI is not introduced in this review and is described thoroughly in many articles [[5][6]]. Briefly, thin sample sections are attached to an electrically conductive steel plate or to an indium-tin oxide (ITO)-coated glass slide immediately followed by matrix application. The sample is then shot with a laser (UV or IR wavelength) whereby the energy is absorbed by the matrix and transferred to the analyte to induce its ionization (Figure 1B).
Table 2. Examples of matrix used in MALDI-MSI.
|
Matrices |
Analytes |
|
N-(1-naphthyl)ethylenediamine dihydrochloride (NEDC) |
glucose |
|
3-amino-4-hydroxybenzoic acid (AHBA) |
glycan |
|
3-aminoquinoline (3AQ) |
glycan |
|
2-,4-,6-trihydroxyacetophenone (THAP) |
glycolipids |
|
N,N-dimethylaniline (DMA) |
hemicellulose |
|
1,5-diaminonapthalene (DAN) |
lipids |
|
2-mercaptobenzothiazole (MBT) |
lipids |
|
dihydroxyacetone phosphate (DHAP) |
lipids, glycan |
|
2,5-dihydroxybenzoic acid (DHB) |
lipids, glycopeptide, polymer |
|
9-aminoacridine (9-AA) |
lipids, metabolites |
|
3-hydroxypicolinic acid (3HPA) |
nucleotides |
|
anthranilic acid (ANA) |
nucleotides |
|
nicotinic acid (NA) |
nucleotides |
|
picolinic acid (PA) |
nucleotides |
|
sinapinic acid (SA) |
peptides, proteins |
|
α-cyano-4-hydroxycinnamic acid (CHCA) |
peptides, proteins |
|
2-(4-hydroxyphenylazo)-benzoic acid (HABA) |
polymer |
|
5-chlorosalycic acid (5CSA) |
polymer |
|
trans-3-indolacrylic acid (IAA) |
polymer, aromatic |
|
nifedipine |
polyphenols |
The newest ionization method among the three mentioned above is DESI. MS analysis by DESI was first reported in 2004 by Takats et al. [[21]]. Unlike MALDI-MS, which is always performed in vacuum, DESI-MS is operated in an ambient atmosphere (Table 1). Electrospray ionization (ESI) directs a pneumatically assisted stream of charged solvent droplets toward the surface of samples, where compounds are extracted by the solvent. Then, the generated secondary ions are propelled into the mass analyzer (Figure 1B). Thus, DESI combines the advantages of ESI and other desorption ionization methods without matrix spraying. This results in minimal damage to the samples without complicated ionization interference by the matrix, which allows the detection of minor components in samples. Other advantages of DESI-MSI is that the samples can be simultaneously applied to both ion imaging and histochemical analyses. Although the lateral resolution of DESI-MSI is modest (50–200 µm) compared with that of other methods, its atmospheric operation allows for the analysis of a wider variety of samples (solid, liquid, frozen, and raw) and does not require sample preparation (Table 1). DESI-MSI has been applied for the characterization of lipids, peptides, and small molecule metabolites in food. Compared with MALDI, currently, along DESI and SIMS-MSI have been less utilized in food science. However, owing to its obvious advantages, DESI-MSI could be widely used in this field.
While SIMS, MALDI, and DESI-MS continue to be widely applied in MSI, new ionization methods are being developed for use in food science. MALDI-MSI has been the most widely used method, however, the necessity of using organic matrices is a disadvantage as described above. Recently, nanoparticles (NPs), including silicones, graphite, and metallic NPs, have been used as an alternative to organic matrices in laser desorption/ionization (LDI)-MSI [[22]], and this technique is called nanoparticle-based surface assisted laser desorption/ionization mass spectrometry imaging (NPs-ALDI-MSI). There are several types of NP-based methods, including silicon (DIOS)-MSI [[23]], and particles containing gold (Au nanoparticle-enhanced target (AuNPET)) [[24][25][26][27]], silver (Ag nanoparticle-enhanced target (AgNPET)) [[27][28]], colloidal graphite (GALDI) [[29]], platinum, copper, nickel, and titanium [[27]] have been used as NPs in MSI research. Many parameters such as composition, particle size, morphology, surface properties, and concentration of nanoparticles affect the performance of NPs-ALDI-MSI. Practical methodologies for ionization using organic matrices and nanoparticles are under discussion [[30]]. It is very difficult to find a general methodology suitable for ionization of all nanoparticles because the ternary interactions of the analyte-nanoparticle-laser on the laser spot are very complex and difficult to study due to the high vacuum within the system. The mechanism of ionization is still a matter of debate, but NPs provide a greater detection sensitivity, wider applications, clear background spectra, and low fragmentation [[22]]. The development and application of new NPs is expected to advance food science research in the future.
3. Future outlook
MSI is a promising technology to visualize the distribution of food components and identify various food factors based on their molecular weights for improved quality, quality assurance, food safety, nutritional analysis, and to locate administered food factors. There are increasing number of studies using MSI technologies in food science and novel technologies are being developing rapidly. For MSI analysis in food science, MALDI is the most commonly used ionization technology. Over the past five years, new organic matrices have been developed for compounds whose localization is desired for analyses (Table 2). New organic matrices continue to be developed and will help advance food science. Recently, NPs based on metals and silicon have been used as a matrix instead of conventional organic matrices. Many parameters affect the performance of NPs, and investigations aimed at establishing the appropriate conditions for the use of NPs will advance food science through the visualization of various molecules in food. Although identification of substances with the same mass (e.g., glucose and fructose) is not possible with current MSI technologies, the problem may be solved with the development of specific enzymatic pretreatment methods and specific inducible reagents. We anticipate that the knowledge obtained from MSI-based technologies in food science along with various other newly developed techniques will soon be widely applicable to other fields of science to aid the study of analytes. However, the detection of compounds that are present at low concentrations or with significantly low ionization efficiency remains a challenge that must be overcome through further study. Therefore, the development of further techniques is necessary to analyze localization of such compounds.
References
- Shin Hye Kim; Jeongkwon Kim; Young Jin Lee; Tae Geol Lee; Sohee Yoon; Sample Preparation of Corn Seed Tissue to Prevent Analyte Relocations for Mass Spectrometry Imaging. Journal of the American Society for Mass Spectrometry 2017, 28, 1729-1732, 10.1007/s13361-017-1682-3.
- Alexandre Seyer; Jacques Einhorn; Alain Brunelle; Olivier Laprévote; Localization of Flavonoids in Seeds by Cluster Time-of-Flight Secondary Ion Mass Spectrometry Imaging. Analytical Chemistry 2010, 82, 2326-2333, 10.1021/ac902528t.
- Katie L. Moore; Markus Schröder; Enzo Lombi; Fang-Jie Zhao; Steve P. McGrath; Malcolm J Hawkesford; Peter R. Shewry; C.R.M. Grovenor; NanoSIMS analysis of arsenic and selenium in cereal grain.. New Phytologist 2009, 185, , 10.1111/j.1469-8137.2009.03071.x.
- Katie L. Moore; Yi Chen; Allison M. L. Van De Meene; Louise Hughes; Wenju Liu; Tina Geraki; J. Frederick W. Mosselmans; Steve P. McGrath; Chris Grovenor; Fang-Jie Zhao; Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytologist 2013, 201, 104-115, 10.1111/nph.12497.
- Yukihiro Yoshimura; Naoko Goto-Inoue; Tatsuya Moriyama; Nobuhiro Zaima; Significant advancement of mass spectrometry imaging for food chemistry. Food Chemistry 2016, 210, 200-211, 10.1016/j.foodchem.2016.04.096.
- Mizuki Morisasa; Tomohiko Sato; Keisuke Kimura; Tsukasa Mori; Naoko Goto-Inoue; Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods 2019, 8, 633, 10.3390/foods8120633.
- Nethaji Gallage; Kirsten Jørgensen; Christian Janfelt; Agnieszka J Z Nielsen; Thomas Naake; Eryk Duński; Lene Dalsten; Michel Grisoni; Birger Lindberg Møller; The Intracellular Localization of the Vanillin Biosynthetic Machinery in Pods of Vanilla planifolia.. Plant And Cell Physiology 2018, 59, 304-318, 10.1093/pcp/pcx185.
- Patel Mohana Kumara; Amitava Srimany; Suganya Arunan; Gudasalamani Ravikanth; Ramanan Uma Shaanker; Thalappil Pradeep; Desorption Electrospray Ionization (DESI) Mass Spectrometric Imaging of the Distribution of Rohitukine in the Seedling of Dysoxylum binectariferum Hook. F. PLOS ONE 2016, 11, e0158099, 10.1371/journal.pone.0158099.
- Paige Hinners; Kelly C. O’Neill; Young Jin Lee; Revealing Individual Lifestyles through Mass Spectrometry Imaging of Chemical Compounds in Fingerprints.. Scientific Reports 2018, 8, 5149, 10.1038/s41598-018-23544-7.
- A Gouaze; J Castaing; J H Soutoul; G Chantepie; [On the orientation of the scapula and of its glenoid cavity].. Archives d'anatomie pathologique 1962, 10, , null.
- Paola Agüi-Gonzalez; Sebastian Jähne; Nhu T. N. Phan; SIMS imaging in neurobiology and cell biology. Journal of Analytical Atomic Spectrometry 2019, 34, 1355-1368, 10.1039/c9ja00118b.
- Richard Caprioli; Imaging Mass Spectrometry: Enabling a New Age of Discovery in Biology and Medicine Through Molecular Microscopy. Journal of the American Society for Mass Spectrometry 2015, 26, 850-852, 10.1007/s13361-015-1108-z.
- Sandra Schulz; Michael Becker; Mark Reid Groseclose; Simone Schadt; Carsten Hopf; Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Current Opinion in Biotechnology 2019, 55, 51-59, 10.1016/j.copbio.2018.08.003.
- Stephanie T. P. Mezger; Alma M. A. Mingels; Otto Bekers; Berta Cillero-Pastor; Ron M. A. Heeren; Trends in mass spectrometry imaging for cardiovascular diseases. Zeitschrift für Analytische Chemie 2019, 411, 3709-3720, 10.1007/s00216-019-01780-8.
- Fei‐Ying Yang; Jun‐Hui Chen; Qian‐Qian Ruan; Hafiz S. A. Saqib; Wei‐Yi He; Min‐Sheng You; Mass spectrometry imaging: An emerging technology for the analysis of metabolites in insects. Archives of Insect Biochemistry and Physiology 2019, 103, e21643, 10.1002/arch.21643.
- Michael. Karas; Franz. Hillenkamp; Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Analytical Chemistry 1988, 60, 2299-2301, 10.1021/ac00171a028.
- Richard Caprioli; Terry B. Farmer; Jocelyn Gile; Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS.. Analytical Chemistry 1997, 69, 4751-4760, 10.1021/ac970888i.
- Joseph A. Hankin; Robert M. Barkley; Robert C. Murphy; Sublimation as a method of matrix application for mass spectrometric imaging. Journal of the American Society for Mass Spectrometry 2007, 18, 1646-1652, 10.1016/j.jasms.2007.06.010.
- Erin Gemperline; Stephanie Rawson; Lingjun Li; Optimization and Comparison of Multiple MALDI Matrix Application Methods for Small Molecule Mass Spectrometric Imaging. Analytical Chemistry 2014, 86, 10030-10035, 10.1021/ac5028534.
- Nobuhiro Zaima; Takahiro Hayasaka; Mitsutoshi Setou; Naoko Goto-Inoue; Application of imaging mass spectrometry for the analysis of Oryza sativa rice. Rapid Communications in Mass Spectrometry 2010, 24, 2723-2729, 10.1002/rcm.4693.
- Zoltan Takats; Justin M. Wiseman; Bogdan Gologan; R. Graham Cooks; Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science 2004, 306, 471-473, 10.1126/science.1104404.
- Hani NasserAbdelhamidab; Nanoparticle-based surface assisted laser desorption ionization mass spectrometry: a review. Mikrochimica Acta 2019, 186, 682, 10.1007/s00604-019-3770-5.
- David Rudd; Kirsten Benkendorff; Charndeep Chahal; Taryn Guinan; Ove Johan Ragnar Gustafsson; Babak Esmaeelian; Hanna Krysinska; Lisa Pogson; Nicolas H. Voelcker; Catherine A. Abbott; Mapping insoluble indole metabolites in the gastrointestinal environment of a murine colorectal cancer model using desorption/ionisation on porous silicon imaging.. Scientific Reports 2019, 9, 12342-13, 10.1038/s41598-019-48533-2.
- Joanna Nizioł; Justyna Sekuła; Tomasz Ruman; Visualizing spatial distribution of small molecules in the rhubarb stalk ( Rheum rhabarbarum ) by surface-transfer mass spectrometry imaging. Phytochemistry 2017, 139, 72-80, 10.1016/j.phytochem.2017.04.006.
- Maria Misiorek; Justyna Sekuła; Tomasz Ruman; Mass Spectrometry Imaging of low Molecular Weight Compounds in Garlic (Allium sativumL.) with Gold Nanoparticle Enhanced Target. Phytochemical Analysis 2017, 28, 479-486, 10.1002/pca.2696.
- Maria Emilia Dueñas; Evan A. Larson; Young Jin Lee; Toward Mass Spectrometry Imaging in the Metabolomics Scale: Increasing Metabolic Coverage Through Multiple On-Tissue Chemical Modifications.. Frontiers in Plant Science 2019, 10, 860, 10.3389/fpls.2019.00860.
- Rebecca L. Hansen; Maria Emilia Dueñas; Young Jin Lee; Sputter-Coated Metal Screening for Small Molecule Analysis and High-Spatial Resolution Imaging in Laser Desorption Ionization Mass Spectrometry. Journal of the American Society for Mass Spectrometry 2018, 30, 299-308, 10.1007/s13361-018-2081-0.
- Joanna Nizioł; Maria Misiorek; Tomasz Ruman; Mass spectrometry imaging of low molecular weight metabolites in strawberry fruit (Fragaria x ananassa Duch.) cv. Primoris with 109Ag nanoparticle enhanced target. Phytochemistry 2019, 159, 11-19, 10.1016/j.phytochem.2018.11.014.
- Sangwon Cha; Hui Zhang; Hilal I. Ilarslan; Eve Syrkin Wurtele; Libuse Brachova; Basil J. Nikolau; Edward S. Yeung; Direct profiling and imaging of plant metabolites in intact tissues by using colloidal graphite-assisted laser desorption ionization mass spectrometry. The Plant Journal 2008, 55, 348-360, 10.1111/j.1365-313x.2008.03507.x.
- Knochenmuss, R. MALDI and Related Methods: A Solved Problem or Still a Mystery? Mass spectrometry 2013, 2, S0006.
- Mizuki Morisasa; Tomohiko Sato; Keisuke Kimura; Tsukasa Mori; Naoko Goto-Inoue; Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods 2019, 8, 633, 10.3390/foods8120633.
- Richard Caprioli; Imaging Mass Spectrometry: Enabling a New Age of Discovery in Biology and Medicine Through Molecular Microscopy. Journal of the American Society for Mass Spectrometry 2015, 26, 850-852, 10.1007/s13361-015-1108-z.





