
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Raimondas Mozūraitis | + 2582 word(s) | 2582 | 2021-07-08 09:50:13 | | | |
| 2 | Amina Yu | + 1 word(s) | 2583 | 2021-07-08 11:30:30 | | | | |
| 3 | Amina Yu | + 1 word(s) | 2583 | 2021-07-08 11:36:13 | | |
Video Upload Options
Sour cherries (Prunus cerasus L.) and sweet cherries (P. avium L.) are economically important fruits with high potential in the food industry and medicine. In this study, we analyzed fungal communities associated with the carposphere of sour and sweet cherries that were freshly harvested from private plantations and purchased in a food store. Following DNA isolation, a DNA fragment of the ITS2 rRNA gene region of each sample was individually amplified and subjected to high-throughput NGS sequencing. Analysis of 168,933 high-quality reads showed the presence of 690 fungal taxa. Investigation of microbial ASVs diversity revealed plant-dependent and postharvest handling-affected fungal assemblages. Among the microorganisms inhabiting tested berries, potentially beneficial or pathogenic fungi were documented. Numerous cultivable yeasts were isolated from the surface of tested berries and characterized by their antagonistic activity. Some of the isolates, identified as Aureobasidium pullulans, Metschnikowia fructicola, and M. pulcherrima, displayed pronounced activity against potential fungal pathogens and showed attractiveness for disease control.
1. Introduction
Sour cherries (Prunus cerasus L.) and sweet cherries (P. avium L.) belong to the family of the Rosaceae [1] and are economically and agronomically important crops. These species are cultivated in temperate and cool regions and characterized by a scattered distribution from the Black Sea to Ireland and Spain, and from Scandinavia to Africa [2]. Cherry fruits are a nutrient-dense food with relatively low caloric content, important nutrients, and significant amounts of bioactive food components, having positive effects on human health [3].
Sour cherry is cultivated for its sharp and juicy fruits that are mostly destined to produce foods like jam, jelly, and syrup or alcohol beverages such as wine, brandy, and fruit beer [1][4]. The current research suggests that intake of fresh sour cherry berries or juice promotes health due to the positive action of anthocyanins and phenolic compounds [3][5]. The consumption of cherries may reduce the risk of arthritis, cancer, cardiovascular disease, and diabetes, and may improve sleep and cognitive functions [3].
Sweet cherry is one of the most economically important fruit species in the world. Most sweet cherry fruits are consumed fresh or processed as frozen, dried, or juiced [3]. Sweet cherry berries are rich in bioactive compounds, including flavonoids, anthocyanins, carotenoids, vitamins, ascorbic acid, and potassium [6] Both laboratory assays and clinical trials have demonstrated the anti-inflammatory, antimicrobial, and anticarcinogenic properties of sweet cherry fruits and their metabolites [6][7][8].
The plant carposphere is highly colonized by various bacterial and fungal microorganisms, the distribution of which is affected by geographic location, climatic conditions, plant species, ripening stage, and growing methods [9][10][11]. Some epiphytic plant-associated microorganisms demonstrate beneficial features, produce secondary metabolites improving resistance in the plant, and impact the structure of the microbial population [12][13][14]. On the other hand, some fruit-inhabiting microorganisms are recognized as pathogenic to hosting plants and humans and are responsible for significant economic losses and serious health problems [15][16]. Therefore, postharvest handling and processing of fruits encounter exceptional attention due to the control of microbiological hazards [17][18].
The scarcity of works characterizing sweet and sour cherries microbial communities are of certain importance. Based on high-throughput sequencing of the 16S rRNA gene, the nitrogen-fixing bacterial community inhabiting Prunus avium L. leaves was described [19] and bacterial diversity of pitted sweet cherries, proceeded high-hydrostatic pressure processing, was evaluated [20]. Endophytic fungi associated with Prunus avium and Prunus cerasus trees located in Germany and Iran were isolated and examined by morphological and molecular analysis [21][22]. Microorganisms present on different cultivars of sour cherries in Hungary were evaluated using cultivation techniques only [18]. It was demonstrated that the frequency of distribution of bacteria, molds, and yeasts were very similar in spite of different cultivars, growing methods, and growing years [18]. To the best of our knowledge, there is no information on sweet and sour cherries carposphere-associated fungal microorganism communities.
2. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts
2.1. Abundance and Diversity of Fungal Microbiota on P. cerasus and P. avium
| Reads Obtained |
HQ Reads |
Observed ASVs |
Pielou Evenness |
Shannon Diversity |
Simpson Index |
|
|---|---|---|---|---|---|---|
| PC-PL1 | 26,895 | 17,126 | 99 | 0.63 | 4.2 | 0.86 |
| PC-PL2 | 28,665 | 19,082 | 109 | 0.66 | 4.46 | 0.91 |
| PC-PL3 | 30,732 | 21,140 | 109 | 0.65 | 4.37 | 0.92 |
| PA-PL1 | 27,571 | 20,325 | 71 | 0.45 | 2.75 | 0.72 |
| PA-PL2 | 28,473 | 17,509 | 52 | 0.54 | 3.08 | 0.78 |
| PA-PL3 | 29,021 | 19,901 | 71 | 0.58 | 3.59 | 0.86 |
| PC-FS1 | 27,825 | 17,104 | 46 | 0.65 | 3.58 | 0.85 |
| PC-FS2 | 30,369 | 19,242 | 58 | 0.65 | 3.78 | 0.87 |
| PC-FS3 | 28,433 | 17,504 | 75 | 0.68 | 4.26 | 0.92 |
| Total: | 257,984 | 168,933 | 690 |
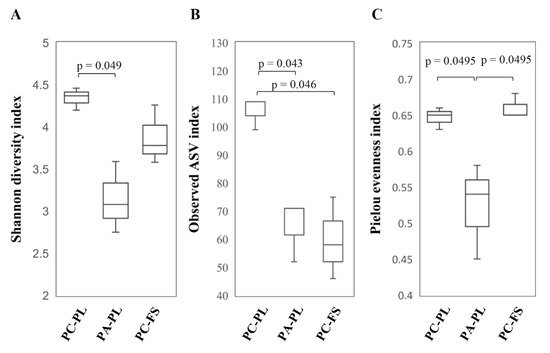
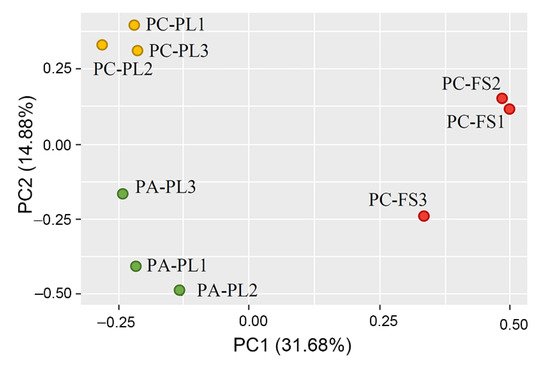
2.2. Fungal Community Profiling
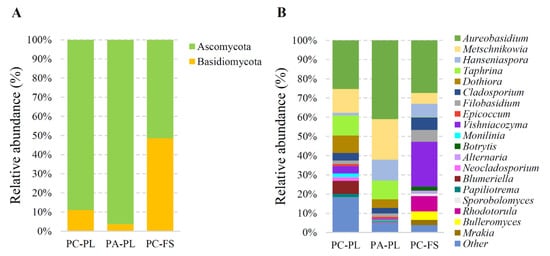
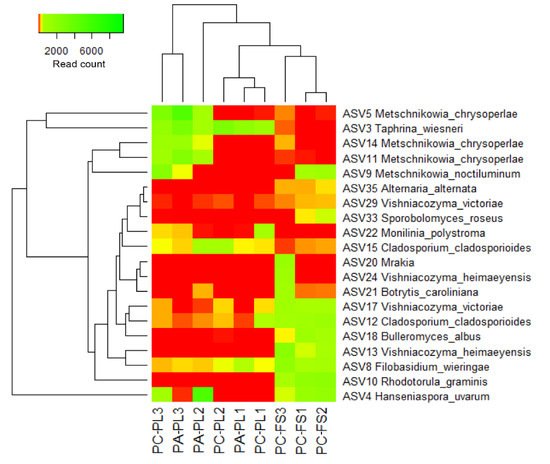
2.3. Distribution of Cultivable Yeasts on P. avium and P. cerasus
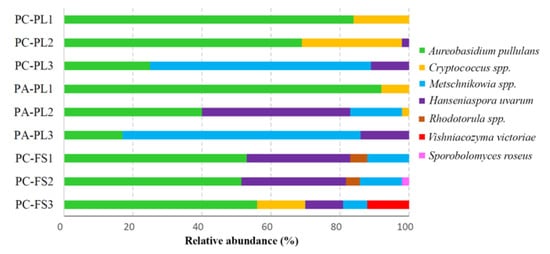
2.4. Antagonistic Activity of Sour and Sweet Cherries-Associated Yeasts
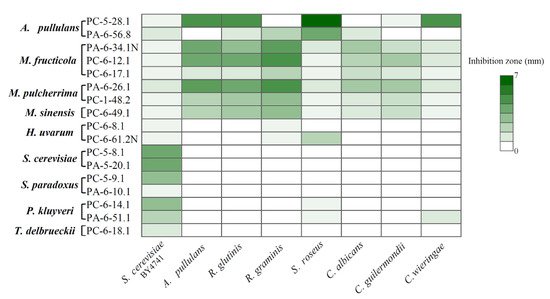
3. Conclusions
References
- Martorell, P.; Fernández-Espinar, M.T.; Querol, A. Molecular Monitoring of Spoilage Yeasts during the Production of Candied FruitNougats to Determine Food Contamination Sources. Int. J. Food Microbiol. 2005, 101, 293–302.
- Bernal-Martinez, L.; Gomez-Lopez, A.; Castelli, M.V.; Mesa-Arango, A.C.; Zaragoza, O.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Susceptibility Profile of Clinical Isolates of Non-Cryptococcus neoformans/Non-Cryptococcus gattii Cryptococcus Species and Literature Review. Med. Mycol. 2010, 48, 90–96.
- McNicholas, S.; McDermott, H.; Power, L.; Johnson, E.M.; Moroney, J.; Humphreys, H.; Smyth, E.G. Sporobolomyces roseus in the Cerebrospinal Fluid of an Immunocompetent Patient--toTreat or Not to Treat? J. Med. Microbiol. 2012, 61, 295–296.
- Liu, D.; Ma, L.; Shi, Y.; Wang, A.; Liu, C. Molecular Diagnosis and Source Tracing of an Infection of Aureobasidium pullulans. J. Infect. Dev. Ctries. 2019, 13, 1174–1179.
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida Albicans-The Virulence Factors and Clinical Manifestations of Infection. J. Fungi 2021, 7, 79.
- Wirth, F.; Goldani, L.Z. Epidemiology of Rhodotorula: An Emerging Pathogen. Interdiscip. Perspect. Infect. Dis. 2012, 465717.
- Zhang, H.; Li, Q.; Qiao, G.; Qiu, Z.; Wen, Z.; Wen, X. Optimizing the Supercritical Carbon Dioxide Extraction of Sweet Cherry (Prunus avium L.) Leaves and UPLC-MS/MS Analysis. Anal. Methods 2020, 12, 3004–3013. [Google Scholar] [CrossRef]
- Fazzari, M.; Fukumoto, L.; Mazza, G.; Livrea, M.A.; Tesoriere, L.; Di Marco, L. In Vitro Bioavailability of Phenolic Compounds from Five Cultivars of Frozen SweetCherries (Prunus avium L.). J. Agric. Food Chem. 2008, 56, 3561–3568. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine Fermentation Microbiome: A Landscape from Different Portuguese WineAppellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef]
- Martins, G.; Miot-Sertier, C.; Lauga, B.; Claisse, O.; Lonvaud-Funel, A.; Soulas, G.; Masneuf-Pomarède, I. Grape Berry Bacterial Microbiota: Impact of the Ripening Process and the FarmingSystem. Int. J. Food Microbiol. 2012, 158, 93–100. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Jurick, W.M., 2nd; Peter, K.A.; Kurtzman, C.P.; Buyer, J.S. Yeasts Associated with Plums and Their Potential for Controlling Brown Rot afterHarvest. Yeast 2014, 31, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol Yeasts: Mechanisms and Applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Vepštaitė-Monstavičė, I.; Apšegaitė, V.; Blažytė-Čereškienė, L.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Ravoitytė, B.; Aleknavičius, D.; Būda, V.; Mozūraitis, R.; et al. Fungal Microbiota of Sea Buckthorn Berries at Two Ripening Stages and VolatileProfiling of Potential Biocontrol Yeasts. Microorganisms 2020, 8, 456. [Google Scholar] [CrossRef]
- Coutinho, T.A.; Venter, S.N. Pantoea Ananatis: An Unconventional Plant Pathogen. Mol. Plant Pathol. 2009, 10, 325–335. [Google Scholar] [CrossRef]
- Lukša, J.; Vepštaitė-Monstavičė, I.; Yurchenko, V.; Serva, S.; Servienė, E. High Content Analysis of Sea Buckthorn, Black Chokeberry, Red and White CurrantsMicrobiota—A Pilot Study. Food Res. Int. 2018, 111, 597–606. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Wisniewski, M.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Metagenomic Analysis of Fungal Diversity on Strawberry Plants and the Effect of Management Practices on the Fungal Community Structure of Aerial Organs. PLoS ONE 2016, 11, e0160470. [Google Scholar] [CrossRef]
- Pintér, S.; Bata-Vidács, I.; Beczner, J. Epiphytic Microbiota of Sour Cherry (Prunus cerasus L.) in Integrated and Organic Growing. Acta Aliment. 2013, 42, 618–630. [Google Scholar] [CrossRef]
- Liang, S.; Liu, H.; Wu, S.; Xu, S.; Jin, D.; Faiola, F.; Zhuang, X.; Zhuang, G.; Qu, D.; Fan, H.; et al. Genetic Diversity of Diazotrophs and Total Bacteria in the Phyllosphere of Pyrus serotina, Prunus armeniaca, Prunus avium, and Vitis vinifera. Can. J. Microbiol. 2019, 65, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Toledo Del Árbol, J.; Pérez Pulido, R.; La Storia, A.; Grande Burgos, M.J.; Lucas, R.; Ercolini, D.; Gálvez, A. Microbial Diversity in Pitted Sweet Cherries (Prunus Avium L.) as Affected byHigh-Hydrostatic Pressure Treatment. Food Res. Int. 2016, 89, 790–796. [Google Scholar] [CrossRef]
- Abdollahi Aghdam, S.; Fotouhifar, K.-B. New Reports of Endophytic Fungi Associated with Cherry (Prunus avium) and Sour Cherry (Prunus cerasus) Trees in Iran. Mycol. Iran. 2016, 3, 75–85. [Google Scholar] [CrossRef]
- Bien, S.; Damm, U. Prunus Trees in Germany—A Hideout of Unknown Fungi? Mycol. Prog. 2020, 19, 667–690. [Google Scholar] [CrossRef]




