Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muntazir Mushtaq | + 2472 word(s) | 2472 | 2021-06-08 13:31:11 | | | |
| 2 | Ron Wang | Meta information modification | 2472 | 2021-06-23 09:47:32 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mushtaq, M. CRISPR-Based Genome Editing Tools. Encyclopedia. Available online: https://encyclopedia.pub/entry/11027 (accessed on 07 February 2026).
Mushtaq M. CRISPR-Based Genome Editing Tools. Encyclopedia. Available at: https://encyclopedia.pub/entry/11027. Accessed February 07, 2026.
Mushtaq, Muntazir. "CRISPR-Based Genome Editing Tools" Encyclopedia, https://encyclopedia.pub/entry/11027 (accessed February 07, 2026).
Mushtaq, M. (2021, June 18). CRISPR-Based Genome Editing Tools. In Encyclopedia. https://encyclopedia.pub/entry/11027
Mushtaq, Muntazir. "CRISPR-Based Genome Editing Tools." Encyclopedia. Web. 18 June, 2021.
Copy Citation
Genome-editing (GE) is having a tremendous influence around the globe in the life science community. Among its versatile uses, the desired modifications of genes, and more importantly the transgene (DNA)-free approach to develop genetically modified organism (GMO), are of special interest. The recent and rapid developments in genome-editing technology have given rise to hopes to achieve global food security in a sustainable manner.
genome editing
CRISPR
base editing
prime editing
DNA-free genome editing
crop improvement
1. Introduction
Keeping in view climate change, feeding of the global population and addressing the concerns of malnutrition, especially in developing countries from the perspective of global population explosion, have become intimidating tasks for the scientific community [1]. The global population could reach 9.6 billion by 2050, which is an increase from the current 7.3 billion [2][3][4][5]. Conventional breeding approaches appear to be insufficient to fulfill the global food demand and other environmental challenges that we face in the 21st century. To meet the current demand, at least 23 percent more agricultural production is needed. In modern agriculture, breeding through mutations, crosses, and transgenics have become the core crop improvement strategies in the present era. However, conventional breeding being entirely dependent on existing genetic variations and crossing strategies to introduce desirable traits into crops means that it takes a considerable amount of time and resources, which will continue to limit crop improvement. Moreover, genetic variation has significantly decreased, preventing the potential of trait improvements through cross breeding. Mutation breeding has introduced genetic variability through random mutations using physical or chemical mutagens [5][6][7][8][9][10]. Moreover, commercialization of GM/biotech crops is limited by public concerns, as well as by long and costly regulatory evaluation processes [4].
Since scientists conducted the first gene-targeting (GT) trial in tobacco protoplasts, and the discovery that gene-targeting efficiency is enhanced via DNA double-strand breaks (DSBs) [11], researchers all over the globe have searched for novel developing tools to edit plant genomes. Genome engineering is revolutionizing plant biology through targeted modification of various regulatory elements or genes, or rearranging chromosomes in elite cultivars. Genome-editing employs various sequence-specific nucleases, called engineered nucleases. It consists of two parts: a DNA targeting part that guides the nuclease to the specific target site in the genome and a nuclease that cuts the genome at specific sites. There are three types of engineered nucleases that have been discovered so far: zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) and CRISPR-Cas system [12][13][14][15][16][17]. These sequence-specific nucleases generate double-stranded breaks (DSBs) at targeted genomic sites, which are in turn repaired by either homology-directed repair (HDR) or the non-homologous end joining (NHEJ) method [17][18][19][20]. Until now, diverse genome editing methods have been used to improve various traits in crop plants (Figure 1) [18][21]. The CRISPR-Cas9 system, which is being widely adopted for plant genome editing, has accelerated the crop breeding beyond what was imaginable before its development. ZFNs and TALENs suffer from technical complexity and low efficiency, but the CRISPR/Cas9 system is simple and highly efficient. Due to its high efficiency, versatility, minimum cost, and ease of execution, the CRISPR/Cas9 has developed into a potential tool for the improvement of several plant genomes, notably major crop species [5][17][18][22][23]. The application of CRISPR/Cas9 in plant breeding using different editing approaches has been illustrated in Figure 2. A comparative table of three engineered nucleases has been made (Table 1).
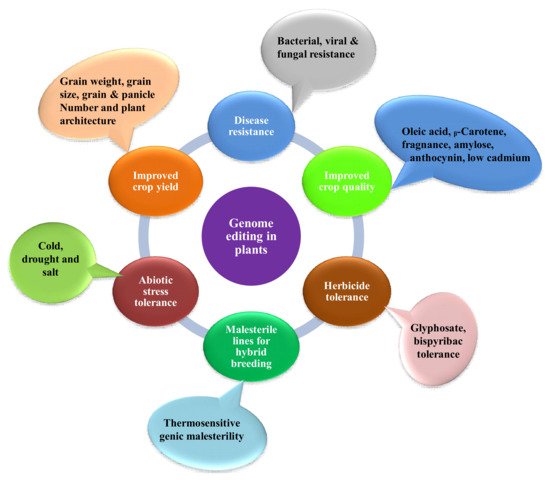
Figure 1. Applications of genome editing in crop plants for improving various traits.
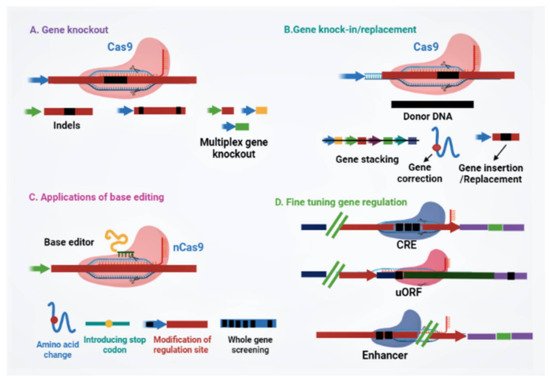
Figure 2. Potential applications of CRISPR/Cas-based applications for plant breeding. (A) CRISPR/Cas-mediated mutation can achieve indels, gene deletions, and multiplex gene knockout. (B) Gene insertion and replacement mediated by either homology-directed repair or non-homologous end joining can accomplish gene stacking for multiple traits, gene correction for gain-of-function, and gene insertion or replacement to generate novel traits for crop improvement. (C) Applications of base editing for crop trait improvement, viz. precise amino acid substitution, gene disruption by introducing a stop codon, gene regulation, and whole-gene screening. (D) CRISPR/Cas system-based gene regulation by engineering the regulatory site in the untranslated region, promoter, or enhancer region. Abbreviations: CRE, cis-regulatory element; sgRNA, single guide RNA; uORF, upstream open reading frame.
Table 1. Comparative table of engineered nucleases.
| S.No. | Properties | ZFN | TALEN | CRISPR-cas9 | References |
|---|---|---|---|---|---|
| 1 | Target site | 20–35bp | 20–40bp | 20–23bp | [24][25][26][27][28] |
| 2 | Nuclease | Two molecules of fokI | Two molecules of fokI | Cas9 | |
| 3 | Efficiency | High | High | High | |
| 4 | Identification molecule | Protein-DNA | Protein-DNA | RNA-DNA hybrid | |
| 5 | Cost | High | Moderate | Low | |
| 6 | Limitation | Time consuming and laborious | Laborious | Off-targeting |
2. Prokaryotic Origin of CRISPR
CRISPR and its related proteins (cas) play very important roles in providing adaptive immunity in prokaryotes and archaea against viruses and plasmids. It relies on the presence of loci in bacteria, called CRISPR, and it was first discovered in E. coli. CRISPR loci consists of operons for the synthesis of the Cas9 protein (nuclease) and repeated spacer sequences. A short fragment (20 bp) of foreign DNA (viral or plasmid) that becomes integrated in bacterial genome is called the protospacer. In the first stage, PAM (protospacer adjacent motif) sequences are identified by bacteria in the invading microbe and then integrates a part of their genome in its CRISPR locus. During the second stage, CRISPR express the entire loci and produces an RNA molecule called crRNA (short for CRISPR-RNA), which, along with Cas9 and other necessary proteins, form a hybrid with invading genomes and Cas9 cuts the genome [29][30][31]. This is how CRISPR works in prokaryotes and helps to protect the bacteria from foreign enemies. This natural system of prokaryotes has been exploited for targeted genome editing in higher organisms, including plants.
3. Working Principle of CRISPR-Cas9 in Plants
Generally, CRISPR-Cas-based editing in plant genomes comprises four steps. First, gene-specific sgRNAs (single guide RNA) are designed using different web-based sgRNA design tools, require that users input a genomic location, gene name or a DNA sequence for each gene of concern, and designate a species. The expression of sgRNA in plants is driven by U3 and U6 small nuclear RNA promoters. In order to enhance the accuracy of computational sgRNA selection, organized studies of sgRNA performances in plant cells and large-scale data collection are needed. The second step involves a transient transformation system like hairy-root or protoplast transformation with CRISPR before being used in genome editing, which is the best way to validate the activity of sgRNA. Thereafter, the third step involves delivery of a genome-editing construct into plant cells, usually via particle bombardment or Agrobacterium-mediated transformation, and a stable integration of sgRNA and Cas9 into the plant genome. In the last step, screening of transformed or regenerated plants for mutation is done via PCR/RE genotyping and is confirmed by sequencing (Figure 3).
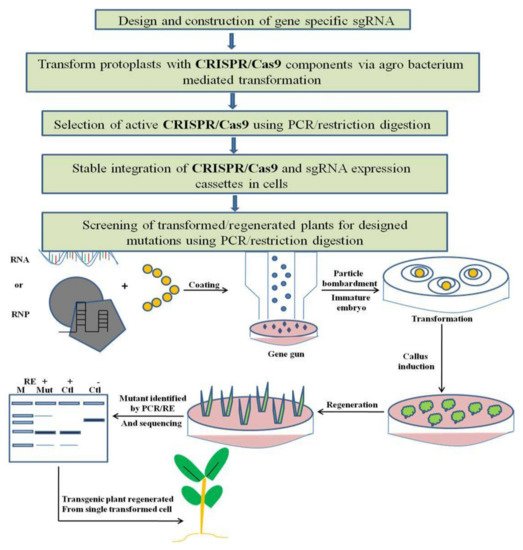
Figure 3. Working principle of CRISPR/Cas9 based genome editing in plants. Plant genome editing can typically be divided into four continuous steps, and the estimated time needed for each step is indicated. PCR/RE, polymerase chain reaction/restriction enzyme digestion. The CRISPR–Cas9 RNAs (in vitro synthesized Cas9 and sgRNA transcripts) or pre-assembled CRISPR–Cas9 RNP can be delivered into immature embryos via particle bombardment. Alternatively, pre-assembled CRISPR–Cas9 RNP can be transfected into plant protoplasts. Bombarded/transfected cells are induced to form calli, from which seedlings are regenerated under the selection-free conditions. Regenerated plants are screened for mutation via the PCR/RE assay and sequencing. Delivering CRISPR–Cas9 reagents via RNP limits their temporal activity, thereby improving their precision. RE, restriction enzyme; M, DNA marker; mut, mutant; ctrl, control.
4. Availability of Compatible Nucleases
Indeed, numerous strategies and tools have been developed to enhance the target specificity of Cas9, including high-fidelity of Cas9 variants and the Cas9 paired nickase strategy [32]. Streptococcus pyrogens SpCas9 is the most common Cas9 used in plants, which recognize the PAM type NGG. While this PAM sequence is widespread across the genomes of plants, it does not cover the whole plant genome. Another problem with SpCas9 is its large protein size (1368 amino acids), which interferes with its delivery through a size limited viral vector, such as the adeno-associated virus (AAV) [33]. PAM sequence requirement restricts the probable target sequences in a gene of interest when it comes to inactivate a gene at any position by targeted mutagenesis. With further studies, the diversity of PAM sequences has been expanded by the novel Cas9 variants and RNA-guided nucleases. Two SpCas9 variants, such as StCas9 [34] from Streptoccocus thermophilus and SaCas9 [34][35] from Staphylococcus aureus, were found to induce targeted plant genome-editing. They might increase the specificity of editing, as both Cas9 variants require longer PAMs.
Furthermore, seven novels programmable CRISPR-Cas nucleases of the Class 2 system were uncovered in bacterial genomes using functional and computational analyses. These comprise five different DNA targeting nucleases (C2c1, C2c3, Cpf1, CasX and CasY) and two other RNA targeting nucleases (C2c2 and C6c6) that belong to type V and VΙ. The role of Cas12a and Cas12b (Cpf1 and C2c1), Cas12c and Cas12d (CasX and CasY), and cas13a (C2c2) in genome engineering was performed in vitro and/or in vivo by various researchers [36]. Differences between these nucleases in terms of nuclease domains, requirement of tracrRNA and crRNA and the DNA cleavage mechanism, were revealed through functional and mechanistic studies [37].
Cas12a, formerly Cpf1 from Prevotella and Francisella, is one of the recently discovered and characterized endonucleases from class II type-V CRISPR-Cas systems, which emerged as the most extensively explored substitute to SpCas9 for plant genome-editing [33]. Cpf1 is capable of non-specific cleavage of ssDNA (single-stranded DNA) and RNA, respectively. Cpf1 with the PAM sequence TTTV where “V” is A, C, or G generates staggered cuts with 5′ overhangs [38][32] in a non-specific manner, which is a completely different way from Cas9 that cleaves dsDNA creating, blunt end DSBs. The former cleavage structure offers certain benefits for directed gene insertion utilizing NHEJ [39]. Cpf1 usually generates larger indels (≥6 nucleotides) besides possessing not only DSB-inducing activity, but also RNase ΙΙΙ activity implicated in pre-crRNA processing [33]. This activity of Cas12a can be exploited in multiplex genome-editing through a tandemly arrayed pre-crRNA-expressing construct [36]. Genome-editing via Cpf1 has been already tested in various plants. Surprisingly, highlighting the high specificity of Cpf1, there was no off-target edits found in these plants [32]. Such novel CRISPR-Cas tools offer potential applications, as well as to tackle specific challenges facing plant genome editing. A group of researchers demonstrated that Cas12a is involved in transcriptional repression in plants; hence, this system, besides being used in genome-editing, can act as an attractive platform for regulating gene expression in plants [40]. Cpf1 may also cleave RNA, which further enhances the functionality to this class of nucleases [41]. Moreover, the capacity of multiplex genome-editing can be improved by the combination of different CRISPR systems [42].
Agricultural production is severely affected by RNA viruses, in contrast to DNA viruses. Cas13a is currently advancing plant biology research and is a promising toolbox in the CRISPR arsenal to contribute to global food protection and do not depend on transgenes. It has been used to combat tobacco RNA viruses and offers a novel technique to confer immunity [43].
5. Breakthrough Technologies in Generating DNA-Free and Genetically Stable CRISPR-Edited Plants
Plant genome-editing involves the transformation of plant cells with the CRISPR/Cas9 construct and the regeneration of whole plants from a few transformed cells. Genome editing has generally been applied to transformable plants, and existing genetic transformation systems are usually specific to the genotype and have been developed in many plant species. Agrobacterium-mediated plant transformation was used extensively as a flexible tool for generating stably transformed model crop plants [44]. The different methods for generating transgene-free and CRISPR-edited plants are illustrated in (Figure 4). Alternatively, Agrobacterium can facilitate transient transformation of plant cells [45][46] and can be used to transiently express Cas9 and gRNA for producing transgene-free genome editing in plants (Figure 4A). This approach has been effectively used to edit the tobacco phytoene desaturase (PDS) gene using the vector system devoid of a selection marker to permit the survival of transiently transformed cells [47] (Figure 4E). In general, 10% of tobacco plants were transgene-free and edited. Moreover, no sexual segregation is required for the removal of transgenes. It is still difficult to detect the desired transgene-free plants using this method. Using Agrobacteria engineered to enhance the production of DNA-free gene edited plants and/or to enhance the transient expression of transgene in plant cells will significantly enhance the efficiency of transgene-free edited plants. Moreover, Agrobacterium is used to transform most plants, but being a plant pathogen raised some regulatory concerns. The potential of plant genome engineering lies within the successful plant transformation. Delivery of CRISPR/Cas9 reagents and plant transformation problems are the major issues of concern. Moreover, the focus should be on novel tools and strategies that unify the approaches across plant species [32].
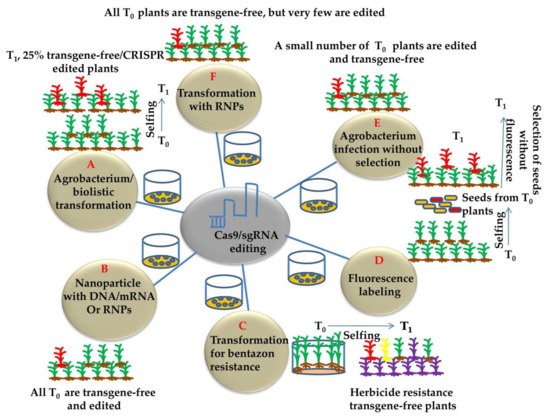
Figure 4. Commonly used approaches for developing transgene-free and CRISPR-edited plants (A) CRISPR gene editing followed by agrobacterium/biolistic transformation obtaining 25% T0 transgene free plants by following Mendelian segregation. (B) Nanoparticle and RNP mediating gene editing. DNA, RNA, or RNP coated nanoparticles can deliver CRIPSR reagents into meristematic cells. This strategy typically produces mosaic plants. The transgene-free and edited plants may be obtained by either sexual or asexual propagation from the edited tissues. (C) Drug-induced elimination of transgenes. The CYP81A6 encodes an enzyme that metabolizes bentazon, which is a herbicide. Coupling CYP81A6 RNAi with CRISPR components enables a selection for transgene-free and edited plants. (D) Fluorescence labeling and selection of transgene free plants. The mCherry fluorescence marker is linked to the gene-editing components in the same plasmid. The marker allows for the selection of transgene-free seeds, greatly reducing the workload associated with growing plants and genotyping. (E) Generation of edited plants using transient expression under no selection pressure. In the absence of a selection pressure, Agrobacterium infection can lead to the expression of transgenes without integrating the transgenes into chromosomes. Such events can lead to the generation of transgene-free and edited plants. (F) Ribonucleoprotein or RNP (Nuclease and gRNA) method generating transgene free plants by particle bombardment/gene gun into calli or immature embryos.
Most plant genome-editing approaches involve the delivery of the expression cassette CRISPR-Cas9 into plant cells, its integration and expression into the nuclear genome and finally the cleavage of the desired nucleotide sequence. However, a small part of dispatched DNA sequence becomes incorporated into the genome. It has also been observed that CRISPR/Cas9 can be transiently expressed and may provide an alternative to plant engineering. Recently, wheat has been transformed via two easy and proficient genome-editing methods that depend on the transient expression of CRISPR-Cas9 RNA or DNA [48]. These two methods removed the selection stage for antibiotics or herbicides in tissue culture during post-transformation stage and callus cells transiently expressing CRISPR-Cas9 were used to regenerate plants. Consequently, tissue culture techniques used were time efficient and less labor intensive. Moreover, transient gene expression systems significantly reduced the transgene integration in wheat callus cells [48]. Genome-editing via CRISPR-Cas9 DNA or RNA transient expression has been established in callus cells of wheat, and can also be successful in other plant species.
References
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754.
- Clarke, J.L.; Zhang, P. Plant biotechnology for food security and bioeconomy. Plant Mol. Biol. 2013, 83, 1–3.
- Haque, E.; Taniguchi, H.; Hassan, M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 617.
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697.
- Zhang, C.; Xu, W.; Wang, F.; Kang, G.; Yuan, S.; Lv, X.; Li, L.; Liu, Y.; Yang, J. Expanding the base editing scope to GA and relaxed NG PAM sites by improved xCas9 system. Plant Biotechnol. J. 2020, 18, 884.
- Adamu, A.K.; Aliyu, H. Morphogical effects of sodium azide on tomato (Lycopersicon esculentum Mill). Sci. World J. 2007, 2, 777–780.
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M. Induced mutagenesis in plants using physical and chemical agents. Plant Cell Cult. Essent. Methods 2010, 20, 111–130.
- Mostafa, G.G. Effect of Sodium Azide on the Grovvth and Variability Induction in. Int. J. Plant Breed. Genet 2011, 5, 76–85.
- Pacher, M.; Puchta, H. From classical mutagenesis to nuclease-based breeding–directing natural DNA repair for a natural end-product. Plant J. 2017, 90, 819–833.
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.M.; Sonah, H. Mutation breeding in tomato: Advances, applicability and challenges. Plants 2019, 8, 128.
- Wright, I.J.; Reich, P.B.; Cornelissen, J.H.C.; Falster, D.S.; Groom, P.K.; Hikosaka, K.; Lee, W.; Lusk, C.H.; Niinemets, Ü.; Oleksyn, J. Modulation of leaf economic traits and trait relationships by climate. Glob. Ecol. Biogeogr. 2005, 14, 411–421.
- Maggio, I.; Goncalves, M.A.F.V. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015, 33, 280–291.
- Mishra, R.; Zhao, K. Genome editing technologies and their applications in crop improvement. Plant Biotechnol. Rep. 2018, 12, 57–68.
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Galla, A.; Abdelrahman, M.; Sharma, M.; Singh, A.K. Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 2019, 10, 550.
- Bao, W.; Wan, Y.; Baluška, F. Nanosheets for delivery of biomolecules into plant cells. Trends Plant Sci. 2017, 22, 445–447.
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674.
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56.
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for crop improvement: An update review. Front. Plant Sci. 2018, 9, 985.
- Puchta, H.; Dujon, B.; Hohn, B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 5055–5060.
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271.
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 2019, 10, 114.
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963.
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52.
- Gaj, T.; Gersbach, C.A.; Barbas Iii, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405.
- Lozano-Juste, J.; Cutler, S.R. Plant genome engineering in full bloom. Trends Plant Sci. 2014, 19, 284–287.
- Jiang, W.Z.; Henry, I.M.; Lynagh, P.G.; Comai, L.; Cahoon, E.B.; Weeks, D.P. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 2017, 15, 648–657.
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Bae, H. Genome editing tools in plants. Genes 2017, 8, 399.
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome editing in plants: Exploration of technological advancements and challenges. Cells 2019, 8, 1386.
- Bannikov, A.V.; Lavrov, A.V. CRISPR/CAS9, the king of genome editing tools. Mol. Biol. 2017, 51, 514–525.
- Karimian, A.; Azizian, K.; Parsian, H.; Rafieian, S.; Shafiei-Irannejad, V.; Kheyrollah, M.; Yousefi, M.; Majidinia, M.; Yousefi, B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. Physiol. 2019, 234, 12267–12277.
- Nussenzweig, P.M.; Marraffini, L.A. Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria. Annu. Rev. Genet. 2020, 54, 93–120.
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 36, 745–757.
- Nakade, S.; Yamamoto, T.; Sakuma, T. Cas9, Cpf1 and C2c1/2/3―What’s next? Bioengineered 2017, 8, 265–273.
- Steinert, J.; Schiml, S.; Fauser, F.; Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 2015, 84, 1295–1305.
- Kaya, H.; Mikami, M.; Endo, A.; Endo, M.; Toki, S. Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 2016, 6, 1–9.
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR crops: Plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 2018, 56, 479–512.
- Murugan, K.; Babu, K.; Sundaresan, R.; Rajan, R.; Sashital, D.G. The revolution continues: Newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell 2017, 68, 15–25.
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771.
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546.
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 1–5.
- Lowder, L.; Malzahn, A.; Qi, Y. Rapid evolution of manifold CRISPR systems for plant genome editing. Front. Plant Sci. 2016, 7, 1683.
- Zhang, D.; Zhang, H.; Li, T.; Chen, K.; Qiu, J.-L.; Gao, C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017, 18, 1–7.
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1–9.
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042.
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-mediated plant transformation: Biology and applications. Arab. Book 2017, 15, e0186.
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693.
- Chen, L.; Li, W.; Katin-Grazzini, L.; Ding, J.; Gu, X.; Li, Y.; Gu, T.; Wang, R.; Lin, X.; Deng, Z. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 2018, 5, 1–12.
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 1–8.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
23 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




