
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Diego La Mendola | + 2431 word(s) | 2431 | 2021-05-13 10:51:51 | | | |
| 2 | Camila Xu | Meta information modification | 2431 | 2021-06-03 08:19:13 | | | | |
| 3 | Camila Xu | Meta information modification | 2431 | 2021-06-10 04:11:54 | | |
Video Upload Options
Nerve growth factor (NGF) is a protein essential to neurons survival, which interacts with its receptor as a non-covalent dimer. Copper affects biological activity of NGF and conversely NGF may regulates copper trafficking in synaptic cleft.
1. Introduction
Copper performs several essential functions as a cofactor in many enzymes in the living systems and it is required for the development and function of the human brain [1][2][3]. In recent years, it has been shown that copper, analogously to zinc, plays a role as modulator of cellular signal transduction pathways [4][5][6]. Copper is stored in synaptic vesicles and is released by electrical depolarization in the synaptic cleft of glutamatergic synapses at concentration values that can reach 100 μM [7]. Copper may have inhibitory or stimulating effect on synaptic plasticity, affecting memory and learning processes although the mechanisms by which the metal performs these functions remain largely undefined [8][9]. The dual effect may be related to the metal binding by different proteins expressed in neurons and released in the synapses as beta-amyloid (Aβ) [10].
The actual biological function of Aβ is still unknown. The polypeptide may control copper efflux in the synapses and it has been demonstrated that the polypeptide improves memory formation [11], synaptic plasticity, and neuronal survival [12].
In particular, the monomeric form of Aβ activates the cyclic AMP response element-binding protein (CREB), which in turn promotes the transcription and release of the brain-derived neurotrophic factor (BDNF) a neurotrophin (NT) strongly involved in long term potentiation (LTP) and memory formation [13].
It is worth noting that it has been demonstrated that copper ions may modulate kinase signaling networks of neuronal tissues induced by neurotrophins [14]. The nerve growth factor (NGF) is the first discovered member of NTs family [15]. NGF is essential for the development, survival, and activity of neurons [16][17]. NGF initiates the signaling pathways through the binding to tropomyosin receptor kinase A (TrkA) triggering a signaling cascade up to the activation of CREB [18][19].
Copper ions enhance NGF functionality and the effect seems related to the presence of metal binding site in the N-terminus domain of NGF [20]. NGF(1-14), a peptide encompassing the first 14 residue of NGF, is able to bind copper ions and mimic whole protein signal transduction activating CREB [21]. The NGF interacts with TrkA as a non-covalent dimer, so a dimeric form of the peptide NGF(1–14) has been tested, showing higher activity than monomers in the release of BDNF [14].
NGF signaling pathways control also the post-translational modifications of the amyloid precursor protein (APP) and then Aβ production in neurons [22][23][24]. Moreover, the deprivation in NGF determines Aβ aggregation and tau hyperphosphorylation in Alzheimer’s disease (AD) [25] while NGF addition protects against cell death and toxicity triggered by Aβ but the underlying mechanism remains unclear [26]. On the other hand, astrocytes activated by Aβ stimulate NGF secretion, whose excess in turn causes the death of hippocampal neurons [27].
In the dynamic environment present at the synaptic cleft, a potential interconnection among copper, Aβ, and NGF, could be one of the key components of the memory formation process.
Therefore, the dyshomeostasis of copper could be at the basis of Aβ and/or NGF dysfunctions; conversely, a malfunction in the metabolic pathways could negatively affect the normal influx of copper into neurons [28]. The consequences could lead to the development of the progressive neurodegenerative disorder such as Alzheimer’s disease (AD) [29][30].
In the brains of patients affected by AD, a high copper concentration, up to 0.4 mM, is localized in senile plaques composed of amyloid aggregates [31]; however, the involvement of copper in AD is still controversial [32]. Studies show copper deficiency in patients affected by AD, and consequently a need to enhance copper levels so to restore normal metal concentration [33][34]; differently, other experiments point to metal overload and consequent demand to reduce copper concentration [35][36].
Copper dyshomeostasis with an increase in the labile pool of metal and a parallel decrease in the copper bound to proteins or peptides is the main interpretation [37][38][39]. A local imbalance of copper between extra- and intra-cellular spaces may drive its binding to Aβ, which may result in formation of oligomeric fibrils, amyloid, or amorphous aggregates, depending on the metal ion/peptide molar ratio.
Aβ may play the role of handling copper cellular influx/efflux via binding metal and transfer it to human copper transporter 1 (hCtr1), the main protein responsible for Cu import into cells [40]. Studies carried out with the peptide model showed that a peptide encompassing the first 14 residues of hCtr1 binds copper ions with higher affinity than N-terminal Aβ peptides [41]. For this reason, it is relevant to know the affinity of Aβ towards copper ions and many studies have been carried out with different techniques to determine the coordination environment of copper complexes formed with Aβ [42][43].
The pH value is another environmental condition that influences Aβ aggregation and Aβ copper binding. The brain pH is imbalanced towards mild acidic condition in aging and patients with AD [44]. A lowering of pH favors Aβ aggregation and strongly alters the release of pro-inflammatory cytokine affecting the uptake of Aβ by microglial cells [45].
Pathologies, such as ischemic stroke, induce a local decrease of extracellular pH due to an inflammatory insult that is as supposed to be an early event in AD progression [46]. Furthermore, the pH influences copper complex speciation with Aβ and mild acidosis can promote conformational changes or reactive oxygen species production promoting Aβ aggregate precipitation [47].
NGF has been postulated as potential therapeutic agent for the treatment of AD and its side effects, as loss of memory, and taking into account that NGF can bind copper ions in the same spaces shared by Aβ, it may be of interest to determine its affinity for copper(II).
Potentiometry is a technique that determines the stability constant values with a certain accuracy and permits also detection of minor species. This experimental technique may be the ideal choice when conditions allow for its application that can be limited by some factors such as poor solubility of longer or hydrophobic peptides. For this reason, potentiometric measurements have been carried out on shorter and more soluble Aβ fragments, in particular the N-terminal ones where the copper binding sites are found [43].
2. Protonation Constants
Peptides protonation constant values were determined by means of potentiometric titrations and are reported in Table 1 together with those of NGF(1-14) peptide. The monomeric ligands rNGF(14-1) and sNGF(1-14) show four proton accepting centers, as expected. The highest pK value corresponds to the N-terminal amino group. The protonation equilibria of the two histidine residues overlap and the average value for the protonation constant values of the imidazole residues is similar to that reported for analogous peptides [48][49]. The lowest pK value belongs to carboxylate side chain of glutamate and it agrees with that found for other peptides containing glutamic acid residues [50].
Table 1. Protonation constant (log βpqr) and pK values (T = 298 °K and I = 0.1 M KNO3) a.
| Species | NGF(1-14) b | rNGF(14-1) | sNGF(1-14) | dNGF(1-15) |
|---|---|---|---|---|
| LH | 7.56 | 7.82 (2) | 7.85 (3) | 7.65 (5) |
| LH2 | 14.13 | 14.51 (2) | 14.65 (3) | 15.35 (2) |
| LH3 | 20.14 | 20.60 (2) | 20.87 (3) | - |
| LH4 | 24.44 | 24.88 (2) | 25.11 (4) | 28.77 (6) |
| LH5 | - | 35.12 (4) | ||
| LH6 | - | 41.31 (4) | ||
| LH7 | - | 46.62 (4) | ||
| LH8 | - | 50.75 (4) | ||
| pK COO- | 4.13 | 4.28 | 4.23 | 4.13 |
| pK COO- | - | - | - | 5.28 |
| pK His | 6.01 | 6.09 | 6.22 | 6.21 |
| pK His | 6.57 | 6.69 | 6.81 | 6.35 |
| pK His (×2) | - | - | - | (6.71 × 2) |
| pK NH2 | 7.56 | 7.82 | 7.85 | 7.65 |
| pK NH2 | - | 7.70 |
a Standard deviations (3σ values) are given in parentheses. b Reference [20].
The dimeric peptide d(NGF1-15) was obtained by disulfide bridge between two units of the sequence 1–15 of the protein, exploiting the cysteine present in position 15. Therefore, the number of protonation sites is double compared to that of monomeric peptides, scrambled and reverse. The pK values of N-terminal groups are very similar to that of monomer NGF(1-14). Protonation reactions of the four imidazole side chains take place in completely overlapping reactions and the measured pK values are in a range between 6.7 and 6.2. The pK value of one carboxylate is the same of monomer peptide NGF(1-14) whereas the other carboxylate is significantly less acidic with a pK of 5.28.
3. Speciation and Characterization of Copper-Peptide Complexes
The stability constants of the Cu2+ complexes are listed in Table 2. The metal ion speciation for each peptide, at 1:1 metal-to-ligand molar ratio, is shown in Figure 1.
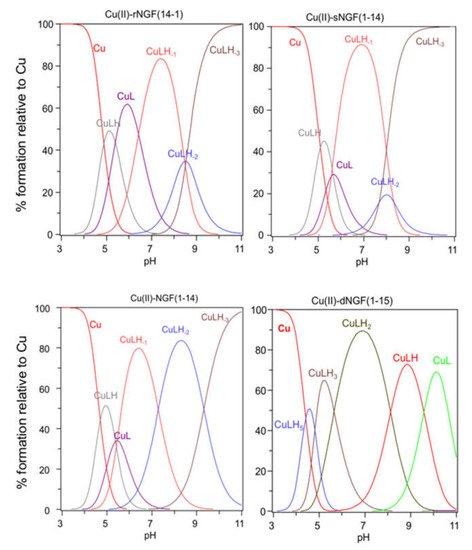
Figure 1. Species distribution of copper(II) complexes with rNGF(14-1), sNGF(1-14), NGF(1-14) and dNGF(1-15). [L] = 1 × 10−3 M; metal to ligand molar ratio of 1:1.
Table 2. Stability constants (log βpqr) and pK values of copper(II) complexes a.
| Species (pqr) b | logβpqr NGF(1-14)c |
logβpqr rNGF(14-1) |
logβpqr sNGF(1-14) |
logβpqr dNGF(1-15) |
|---|---|---|---|---|
| CuLH5 | - | - | - | 41.18 (3) |
| CuLH3 | - | - | - | 31.56 (2) |
| CuLH2 | - | - | - | 25.91 (5) |
| CuLH | 14.09 | 14.15 (1) | 13.97 (4) | 17.79 (5) |
| CuL | 8.72 | 8.77 (1) | 8.32 (5) | 8.21 (4) |
| CuLH-1 | 3.27 | 2.33 (2) | −2.74 (3) | - |
| CuLH-2 | −4.02 | −6.15 (4) | −5.58 (8) | - |
| CuLH-3 | −13.34 | −14.67 (2) | −13.27 (4) | - |
| pK (n/m) | ||||
| pK (5/3) | - | - | - | 4.81 × 2 |
| pK (3/2) | - | - | - | 5.65 |
| pK (2/1) | - | - | - | 8.12 |
| pK (1/0) | 5.37 | 5.38 | 5.65 | 9.57 |
| pK (0/−1) | 5.45 | 6.43 | 5.57 | - |
| pK (−1/−2) | 7.29 | 8.48 | 8.33 | - |
| pK (−2/−3) | 9.30 | 8.52 | 7.69 | - |
a Standard deviations (3σ values) are given in parentheses; b pCu + qH + rL = CupHqLr; βbqr = [CupHqLr]/[Cu]p[H]q[L]r; c Ref. [20]. Charges are omitted for clarity; pK(n/m) values reflect the pK value of copper(II) complexes; [L] = 1 × 10−3 M; molar ratio 1:1.
[CuLH] is the first complex species formed by rNGF(14-1) and sNGF(1-14) and reaches its maximum percentage of formation at pH 5 (Figure 1). The calculated stability constant values (logK(111) = logβ(111) − logβ(011)) are 6.33 and 6.12, for rNGF(14-1) and sNGF(1-14) respectively; the value obtained for reverse sequence is more similar to that of NGF(1-14) (logK(111) = 6.53). The logK(111) values are in the range 6.0–6.5, then higher than those of similar Cu2+ complexes of peptide fragments in which the metal ion is bound to one imidazole nitrogen and one carboxylate [48][51][52]. Indeed, these values are in good agreement with those reported for analogous complex species formed by Aβ peptide fragments for which metal ions have been shown to form a macrochelate with a 2Nim, COO coordination mode (log K = 6.18) [53][54].
Spectroscopic parameters measured at pH = 5, confirm the 2NIm,COO− coordination mode for copper ion and the higher stability of metal complex formed by reverse (λmax = 640 nm, ε = 64 M−1 cm−1) compared to scrambled peptide (λmax = 670 nm, ε = 40 M−1 cm−1) (Table 3). However, it must be underlined that the parameters are partly influenced by the presence of other species other than free copper.
Table 3. Spectroscopic parameters of Copper (II) complexes.
| Peptide | pH | UV-vis λ (nm) (ε (M−1 cm−1)) |
CD λ (nm) (Δε (M−1 cm−1) |
|---|---|---|---|
| rNGF(14-1) | 5 | 640 (64) | 280 (−0.30); 316 (+0.20); 670 (−0.27) |
| 6 | 625 (144) | 280 (−0.40); 323 (+0.33); 671 (−0.44) | |
| 7.4 | 609 (178) | 280 (−0.30); 314 (−0.22); 352 (+0.07); 508 (+0.31); 625 (−0.60) | |
| 9 | 532 (194) | 280 (−1.30); 484 (+0.46); 589 (−0.79) | |
| 10 | 522 (232) | 280 (−1.80); 483 (+0.54); 577 (−1.00) | |
| sNGF(1-14) | 5 | 670 (40) | 289 (−0.20); 328 (+0.12); 647 (−0.04) |
| 6 | 617 (94) | 287 (−1.34); 328 (+0.68); 617 (−0.59) | |
| 7.4 | 603 (102) | 288 (−1.38); 329 (+0.82); 603 (−0.72) | |
| 9-10 | 522 (141) | 306 (+1.47); 544 (−1.13) | |
| dNGF(1-15) | 5 | 635 (50) | 280 (−0.34); 321 (+0.19); 669 (−0.24) |
| 6 | 612 (70) | 280 (−0.40); 323 (+0.33); 671 (−0.44) | |
| 7.4 | 569 (94) | 280 (−1.80); 324 (+0.92); 504 (+0.08); 611 (−0.30) | |
| 8 | 561 (104) | 280 (−2.04); 323 (+0.98); 496 (+0.11); 586 (−0.34) | |
| 9 | 554 (119) | 280 (−2.15); 324 (+0.98); 496 (+0.08); 587 (−0.38) | |
| 10 | 530 (138) | 280 (−2.14); 324 (+0.71); 563 (−0.46) |
Increasing the pH, [CuL] complex species is formed. This species is a minor one but the obtained logβ value suggest the involvement of a further donor atom as N-terminal group for both peptides. The reverse peptide also shows for this species a higher value than scrambled one and very similar to that reported for NGF(1-14).
The contemporary presence of an isomer in which a deprotonated amide is bound to metal instead of an amino group, that remains still protonated, cannot be ruled out.
In particular the Cu-rNGF(14-1) system shows a CD signal centered around 320 nm, that is diagnostic of a charge transfer from a deprotonated amide nitrogen to copper ion [55] (Figure 2).
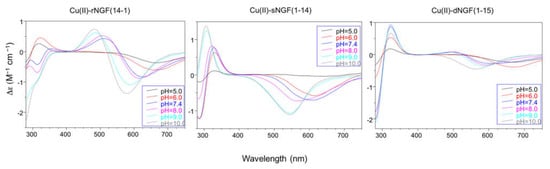
Figure 2. Circular Dichroism spectra with: rNGF(14-1), sNGF(1-14), and dNGF(1-15). [L] = 1 × 10−3 M; metal to ligand molar ratio of 1:1.
The next species formed, [CuLH-1], is the predominant complex in the pH range 6.5–8.0. The stepwise constant values logK(11-1) (logK(11-1) = logβ(110) − logβ(11-1)) are 6.43 for rNGF(14-1) and 5.57 for sNGF(1-14). Both values indicate the deprotonation of an amide nitrogen atom but this step is more favored for sNGF(1-14). The UV–vis parameters are similar suggesting an analogous coordination environment of metal ion with three nitrogen atoms involved in the binding. CD spectra show the diagnostic signal of deprotonated amide nitrogen bound to copper ion even though the conformational features of two peptides are different due to distinctive primary sequence (Figure 2).
As the pH increases, the second deprotonation occurs but it is less favored than the third nitrogen amide deprotonation step. Indeed, the [CuLH-2] complex species is a minor one for both peptides, suggesting that the coordination of successive amide nitrogen deprotonation reactions are accompanied by the rearrangement of the peptide metal binding sites [56]. Namely, the histidine imidazole moiety and the subsequent amide are the primary binding sites below pH 8.5, but they are partly replaced by the amino group and preceding amide functions at higher pH values. This effect is supported by significant blue shift of the absorption spectra as well as of the CD band characteristic of peptide amino-bonded copper(II) complexes [57].
The first species formed by dimeric peptide dNGF(1-15) is [CuLH5] that reaches its maximum percentage at pH 4.5 (Figure 1).
The stability constant (logK(115) = logβ(115) -logβ(015) = 41.18 − 35.12 = 6.06) is indicative of a 2Nim, COO− coordination mode analogous to monomeric peptides and Aβ as above reported. This is further confirmed by UV–vis parameters similar to that of rNGF(14-1) copper complex. It is to note that in the [CuLH5] species there are only three deprotonated centers and therefore the involvement of 2Nim requires that one carboxylate group is still protonated. The unusual high pK value of one carboxylate moiety (pK = 5.28) and the pH range of species existence, between 4.0–5.5, makes this hypothesis plausible.
[CuLH3] is the next species formed with a maximum percentage of formation around pH 5.5 (Figure 1). It is not possible to calculate the stepwise stability constant value as performed for other species because the protonation constant of the [LH3] species was not experimentally obtained. However, the difference with the stability constant of [CuLH5] (logK(112) = logβ(115) − logβ(113) = 9.62) suggests the deprotonation of the carboxylate not coordinated to copper ion and the binding of another nitrogen atom (imidazole or amino) to the metal ion.
[CuLH2] is the predominant species at physiological pH; the stability constant value logK (logK(112) = logβ(112) − logβ(012) = 10.56) is high and can be related with the relevant blue shift in the UV–vis maximum absorption (λmax = 569 nm, ε = 94 M−1 cm−1). All these data are indicative of a strong ligand field around copper ion, determined by four nitrogen atoms coordination mode in a planar arrangement. It can be assumed that the (NH2, N−) five-membered chelate is assisted by the macrochelation with the NIm donor of the His-4 residue of the same chain and the other histidine belonging to the other chain of dimeric peptide.
The CD spectra show an increase in the wide band centered at 324 nm, that include charge transfer signals of both imidazole and deprotonated amide to metal ion, and a d–d transition band with a relatively low intensity compared to the analogous species formed by monomeric peptides. These CD features are in agreement with the involvement of more imidazole side chain in the metal binding (Figure 2).
The next species [CuLH] and [CuL] show similar spectroscopic parameters to those of [CuLH2], suggesting the deprotonation of amino/imidazole not bound to metal ion and afterwards, at strongly basic pH, the deprotonation of an amide nitrogen atom that substitutes one imidazole in the coordination to metal ion.
References
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Metabolism and functions of copper in brain. Prog. Neurobiol. 2014, 116, 33–57.
- Ackerman, C.M.; Chang, C.J. Copper signaling in the brain and beyond. J. Biol. Chem. 2018, 293, 4628–4635.
- Lutsenko, S.; Washington-Hughes, C.; Ralle, M.; Schmidt, K. Copper and the brain noradrenergic system. J. Biol. Inorg. Chem. 2019, 24, 1179–1188.
- Grubman, A.; White, A.R. Copper as a key regulator of cell signalling pathways. Expert Rev. Mol. Med. 2014, 16, e11.
- Kardos, J.; Héja, L.; Simon, Á.; Jablonkai, I.; Kovács, R.; Jemnitz, K. Copper signalling: Causes and consequences. Cell Commun. Signal. 2018, 16.
- La Mendola, D.; Giacomelli, C.; Rizzarelli, E. Intracellular Bioinorganic Chemistry and Cross Talk Among Different -Omics. Curr. Top. Med. Chem. 2016, 16, 3103–3130.
- Kardos, J.; Kovács, I.; Hajós, F.; Kálmán, M.; Simonyi, M. Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability. Neurosci. Lett. 1989, 103, 139–144.
- D’Ambrosi, N.; Rossi, L. Copper at synapse: Release, binding and modulation of neurotransmission. Neurochem. Int. 2015, 90, 36–45.
- Kapkaeva, M.R.; Popova, O.V.; Kondratenko, R.V.; Rogozin, P.D.; Genrikhs, E.E.; Stelmashook, E.V.; Skrebitsky, V.G.; Khaspekov, L.G.; Isaev, N.K. Effects of copper on viability and functional properties of hippocampal neurons in vitro. Exp. Toxicol. Pathol. 2017, 69, 259–264.
- Nam, E.; Nam, G.; Lim, M.H. Synaptic Copper, Amyloid-β, and Neurotransmitters in Alzheimer’s Disease. Biochemistry 2020, 59, 15–17.
- Garcia-Osta, A.; Alberini, C.M. Amyloid beta mediates memory formation. Learn. Mem. 2009, 16, 267–272.
- Parihar, M.S.; Brewer, G.J. Amyloid-β as a modulator of synaptic plasticity. J. Alzheimers Dis. 2010, 22, 741–763.
- Zimbone, S.; Monaco, I.; Gianì, F.; Pandini, G.; Copani, A.G.; Giuffrida, M.L.; Rizzarelli, E. Amyloid Beta monomers regulate cyclic adenosine monophosphate response element binding protein functions by activating type-1 insulin-like growth factor receptors in neuronal cells. Aging Cell 2018, 17, e12684.
- Naletova, I.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Pandini, G.; Triaca, V.; Amadoro, G.; Latina, V.; Calissano, P.; Travaglia, A.; et al. The Copper(II)-Assisted Connection between NGF and BDNF by Means of Nerve Growth Factor-Mimicking Short Peptides. Cells 2019, 8, 301.
- Levi-Montalcini, R. The nerve growth factor 35 years later. Science 1987, 237, 1154–1162.
- Sofroniew, M.V.; Howe, C.L.; Mobley, W.C. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 2001, 24, 1217–1281.
- Conner, J.M.; Franks, K.M.; Titterness, A.K.; Russell, K.; Merrill, D.A.; Christie, B.R.; Sejnowski, T.J.; Tuszynski, M.H. NGF is essential for hippocampal plasticity and learning. J. Neurosci. 2009, 29, 10883–10889.
- Reichardt, L.F. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1545–1564.
- Riccio, A.; Pierchala, B.A.; Ciarallo, C.L.; Ginty, D.D. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science 1997, 277, 1097–1100.
- Travaglia, A.; Arena, G.; Fattorusso, R.; Isernia, C.; La Mendola, D.; Malgieri, G.; Nicoletti, V.G.; Rizzarelli, E. The inorganic perspective of nerve growth factor: Interactions of Cu2+ and Zn2+ with the N-terminus fragment of nerve growth factor encompassing the recognition domain of the TrkA receptor. Chemistry 2011, 17, 3726–3738.
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Side of NGF: Copper(II) and Zinc(II) Affect the NGF Mimicking Signaling of the N-Terminus Peptides Encompassing the Recognition Domain of TrkA Receptor. Front. Neurosci. 2016, 10.
- Mufson, E.J.; Counts, S.E.; Ginsberg, S.D.; Mahady, L.; Perez, S.E.; Massa, S.M.; Longo, F.M.; Ikonomovic, M.D. Nerve Growth Factor Pathobiology during the Progression of Alzheimer’s Disease. Front. Neurosci. 2019, 13.
- Matrone, C.; Ciotti, M.T.; Mercanti, D.; Marolda, R.; Calissano, P. NGF and BDNF signaling control amyloidogenic route and Abeta production in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 13139–13144.
- Canu, N.; Pagano, I.; La Rosa, L.R.; Pellegrino, M.; Ciotti, M.T.; Mercanti, D.; Moretti, F.; Sposato, V.; Triaca, V.; Petrella, C.; et al. Association of TrkA and APP Is Promoted by NGF and Reduced by Cell Death-Promoting Agents. Front. Mol. Neurosci. 2017, 10.
- Canu, N.; Amadoro, G.; Triaca, V.; Latina, V.; Sposato, V.; Corsetti, V.; Severini, C.; Ciotti, M.T.; Calissano, P. The Intersection of NGF/TrkA Signaling and Amyloid Precursor Protein Processing in Alzheimer’s Disease Neuropathology. Int. J. Mol. Sci. 2017, 18, 1319.
- Su, R.; Su, W.; Jiao, Q. NGF protects neuroblastoma cells against β-amyloid-induced apoptosis via the Nrf2/HO-1 pathway. FEBS Open Bio 2019, 9, 2063–2071.
- Sáez, E.T.; Pehar, M.; Vargas, M.R.; Barbeito, L.; Maccioni, R.B. Production of nerve growth factor by beta-amyloid-stimulated astrocytes induces p75NTR-dependent tau hyperphosphorylation in cultured hippocampal neurons. J. Neurosci. Res. 2006, 84, 1098–1106.
- Ejaz, H.W.; Wang, W.; Lang, M. Copper Toxicity Links to Pathogenesis of Alzheimer’s Disease and Therapeutics Approaches. Int. J. Mol. Sci. 2020, 21, 7660.
- Kaden, D.; Bush, A.I.; Danzeisen, R.; Bayer, T.A.; Multhaup, G. Disturbed copper bioavailability in Alzheimer’s disease. Int. J. Alzheimers Dis. 2011, 2011.
- Barnham, K.J.; Bush, A.I. Metals in Alzheimer’s and Parkinson’s diseases. Curr. Opin. Chem. Biol. 2008, 12, 222–228.
- Lovell, M.A.; Robertson, J.D.; Teesdale, W.J.; Campbell, J.L.; Markesbery, W.R. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci. 1998, 158, 47–52.
- Bagheri, S.; Squitti, R.; Haertlé, T.; Siotto, M.; Saboury, A.A. Role of Copper in the Onset of Alzheimer’s Disease Compared to Other Metals. Front. Aging Neurosci. 2018, 9.
- Exley, C.; House, E.; Polwart, A.; Esiri, M.M. Brain burdens of aluminum, iron, and copper and their relationships with amyloid-β pathology in 60 human brains. J. Alzheimers Dis. 2012, 31, 725–730.
- Scholefield, M.; Church, S.J.; Xu, J.; Patassini, S.; Roncaroli, F.; Hooper, N.M.; Unwin, R.D.; Cooper, G.J.S. Widespread Decreases in Cerebral Copper Are Common to Parkinson’s Disease Dementia and Alzheimer’s Disease Dementia. Front. Aging Neurosci. 2021, 13.
- Squitti, R.; Siotto, M.; Polimanti, R. Low-copper diet as a preventive strategy for Alzheimer’s disease. Neurobiol. Aging 2014, 35 (Suppl. 2), S40–S50.
- Behzadfar, L.; Abdollahi, M.; Sabzevari, O.; Hosseini, R.; Salimi, A.; Naserzadeh, P.; Sharifzadeh, M.; Pourahmad, J. Potentiating role of copper on spatial memory deficit induced by beta amyloid and evaluation of mitochondrial function markers in the hippocampus of rats. Metallomics 2017, 9, 969–980.
- Squitti, R.; Ventriglia, M.; Gennarelli, M.; Colabufo, N.A.; El Idrissi, I.G.; Bucossi, S.; Mariani, S.; Rongioletti, M.; Zanetti, O.; Congiu, C.; et al. Non-Ceruloplasmin Copper Distincts Subtypes in Alzheimer’s Disease: A Genetic Study of ATP7B Frequency. Mol. Neurobiol. 2017, 54, 671–681.
- Zhao, J.; Shi, Q.; Tian, H.; Li, Y.; Liu, Y.; Xu, Z.; Robert, A.; Liu, Q.; Meunier, B. TDMQ20, a Specific Copper Chelator, Reduces Memory Impairments in Alzheimer’s Disease Mouse Models. ACS Chem. Neurosci. 2021, 12, 140–149.
- Wang, L.; Yin, Y.L.; Liu, X.Z.; Shen, P.; Zheng, Y.G.; Lan, X.R.; Lu, C.B.; Wang, J.Z. Current understanding of metal ions in the pathogenesis of Alzheimer’s disease. Transl. Neurodegener. 2020, 9.
- Stefaniak, E.; Pushie, M.J.; Vaerewyck, C.; Corcelli, D.; Griggs, C.; Lewis, W.; Kelley, E.; Maloney, N.; Sendzik, M.; Bal, W.; et al. Exploration of the Potential Role for Aβ in Delivery of Extracellular Copper to Ctr1. Inorg. Chem. 2020, 59, 16952–16966.
- Stefaniak, E.; Bal, W. CuII Binding Properties of N-Truncated Aβ Peptides: In Search of Biological Function. Inorg. Chem. 2019, 58, 13561–13577.
- Alies, B.; Bijani, C.; Sayen, S.; Guillon, E.; Faller, P.; Hureau, C. Copper coordination to native N-terminally modified versus full-length amyloid-β: Second-sphere effects determine the species present at physiological pH. Inorg. Chem. 2012, 51, 12988–13000.
- Arena, G.; Pappalardo, G.; Sovago, I.; Rizzarelli, E. Copper(II) interaction with amyloid-β: Affinity and speciation. Coord. Chem. Rev. 2012, 256, 3–12.
- Lyros, E.; Ragoschke-Schumm, A.; Kostopoulos, P.; Sehr, A.; Backens, M.; Kalampokini, S.; Decker, Y.; Lesmeister, M.; Liu, Y.; Reith, W.; et al. Normal brain aging and Alzheimer’s disease are associated with lower cerebral pH: An in vivo histidine 1H-MR spectroscopy study. Neurobiol. Aging 2020, 87, 60–69.
- Decker, Y.; Németh, E.; Schomburg, R.; Chemla, A.; Fülöp, L.; Menger, M.D.; Liu, Y.; Fassbender, K. Decreased pH in the aging brain and Alzheimer’s disease. Neurobiol. Aging 2021, 101, 40–49.
- Tóth, M.O.; Menyhárt, Á.; Frank, R.; Hantosi, D.; Farkas, E.; Bari, F. Tissue Acidosis Associated with Ischemic Stroke to Guide Neuroprotective Drug Delivery. Biology 2020, 9, 460.
- Su, Y.; Chang, P.T. Acidic pH promotes the formation of toxic fibrils from beta-amyloid peptide. Brain Res. 2001, 893, 287–291.
- Grasso, G.; Magrì, A.; Bellia, F.; Pietropaolo, A.; La Mendola, D.; Rizzarelli, E. The copper(II) and zinc(II) coordination mode of HExxH and HxxEH motif in small peptides: The role of carboxylate location and hydrogen bonding network. J. Inorg. Biochem. 2014, 130, 92–102.
- Rajković, S.; Kállay, C.; Serényi, R.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Complex formation processes of terminally protected peptides containing two or three histidyl residues. Characterization of the mixed metal complexes of peptides. Dalton Trans. 2008, 37, 5059–5071.
- Kállay, C.; Várnagy, K.; Micera, G.; Sanna, D.; Sóvágó, I. Copper(II) complexes of oligopeptides containing aspartyl and glutamyl residues. Potentiometric and spectroscopic studies. J. Inorg. Biochem. 2005, 99, 1514–1525.
- La Mendola, D.; Magrì, A.; Campagna, T.; Campitiello, M.A.; Raiola, L.; Isernia, C.; Hansson, O.; Bonomo, R.P.; Rizzarelli, E. A doppel alpha-helix peptide fragment mimics the copper(II) interactions with the whole protein. Chemistry 2010, 16, 6212–6223.
- Karavelas, T.; Malandrinos, G.; Hadjiliadis, N.; Mlynarz, P.; Kozlowski, H.; Barsan, M.; Butler, I. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment -TYTEHA- of histone H4. Dalton Trans. 2008, 9, 1215–1223.
- Kowalik-Jankowska, T.; Ruta, M.; Wiśniewska, K.; Lankiewicz, L. Coordination abilities of the 1-16 and 1-28 fragments of beta-amyloid peptide towards copper(II) ions: A combined potentiometric and spectroscopic study. J. Inorg. Biochem. 2003, 95, 270–282.
- Damante, C.A.; Osz, K.; Nagy, Z.; Pappalardo, G.; Grasso, G.; Impellizzeri, G.; Rizzarelli, E.; Sóvágó, I. The metal loading ability of beta-amyloid N-terminus: A combined potentiometric and spectroscopic study of copper(II) complexes with beta-amyloid(1-16), its short or mutated peptide fragments, and its polyethylene glycol (PEG)-ylated analogue. Inorg. Chem. 2008, 47, 9669–9683.
- Magrì, A.; Munzone, A.; Peana, M.; Medici, S.; Zoroddu, M.A.; Hansson, O.; Satriano, C.; Rizzarelli, E.; La Mendola, D. Coordination Environment of Cu(II) Ions Bound to N-Terminal Peptide Fragments of Angiogenin Protein. Int. J. Mol. Sci. 2016, 17, 1240.
- Kállay, C.; Nagy, Z.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sóvágó, I. Thermodynamic and structural characterization of the copper(II) complexes of peptides containing both histidyl and aspartyl residues. Bioinorg. Chem. Appl. 2007, 2007.
- La Mendola, D.; Magrì, A.; Hansson, Ö.; Bonomo, R.P.; Rizzarelli, E. Copper(II) complexes with peptide fragments encompassing the sequence 122-130 of human doppel protein. J. Inorg. Biochem. 2009, 103, 758–765.




