| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ana Masara Ahmad Mokhtar | + 2619 word(s) | 2619 | 2021-05-19 08:39:31 | | | |
| 2 | Peter Tang | Meta information modification | 2619 | 2021-05-27 03:35:08 | | |
Video Upload Options
As an atypical member of the Rho family small GTPases, RhoH shares less than 50% sequence similarity with other members, and its expression is commonly observed in the haematopoietic lineage. To date, RhoH function was observed in regulating T cell receptor signalling, and less is known in other haematopoietic cells. Its activation may not rely on the standard GDP/GTP cycling of small G proteins and is thought to be constitutively active because critical amino acids involved in GTP hydrolysis are absent. Alternatively, its activation can be regulated by other types of regulation, including lysosomal degradation, somatic mutation and transcriptional repressor, which also results in an altered protein expression. Aberrant protein expression of RhoH has been implicated not only in B cell malignancies but also in immune-related diseases, such as primary immunodeficiencies, systemic lupus erythematosus and psoriasis, wherein its involvement may provide the link between immune-related diseases and cancer.
1. Introduction
|
Rho BTB1 |
Rho BTB2 |
RhoH |
Rnd1 |
Rnd2 |
Rnd3 |
RhoD |
RhoF |
RhoA |
RhoC |
RhoB |
Wrch2 |
Wrch1 |
TC10 |
TCL |
Cdc42 |
RhoG |
Rac2 |
Rac1 |
Rac3 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
RhoBTB1 |
70 |
34 |
33 |
31 |
30 |
34 |
28 |
38 |
37 |
38 |
34 |
32 |
38 |
35 |
40 |
40 |
41 |
42 |
41 |
|
|
RhoBTB2 |
70 |
32 |
32 |
31 |
29 |
34 |
28 |
39 |
37 |
38 |
33 |
32 |
39 |
37 |
40 |
40 |
42 |
42 |
41 |
|
|
RhoH |
34 |
32 |
29 |
32 |
36 |
38 |
33 |
40 |
40 |
41 |
41 |
38 |
40 |
39 |
42 |
40 |
40 |
41 |
40 |
|
|
Rnd1 |
32 |
32 |
29 |
53 |
61 |
37 |
39 |
41 |
42 |
41 |
31 |
32 |
36 |
34 |
37 |
37 |
39 |
39 |
38 |
|
|
Rnd2 |
31 |
31 |
32 |
53 |
63 |
39 |
41 |
46 |
47 |
43 |
28 |
31 |
36 |
35 |
37 |
41 |
40 |
41 |
39 |
|
|
Rnd3 |
29 |
29 |
36 |
61 |
63 |
37 |
40 |
48 |
48 |
47 |
31 |
32 |
39 |
35 |
38 |
41 |
39 |
42 |
40 |
|
|
RhoD |
34 |
35 |
38 |
37 |
39 |
37 |
49 |
49 |
49 |
49 |
39 |
36 |
42 |
38 |
43 |
44 |
46 |
49 |
49 |
|
|
RhoF |
28 |
28 |
33 |
39 |
41 |
40 |
49 |
47 |
48 |
47 |
36 |
37 |
46 |
43 |
43 |
46 |
50 |
59 |
47 |
|
|
RhoA |
38 |
39 |
40 |
41 |
46 |
48 |
49 |
47 |
92 |
85 |
40 |
44 |
51 |
48 |
53 |
55 |
53 |
57 |
55 |
|
|
RhoC |
37 |
37 |
40 |
42 |
47 |
48 |
49 |
48 |
92 |
85 |
40 |
44 |
50 |
49 |
51 |
55 |
53 |
57 |
54 |
|
|
RhoB |
38 |
38 |
41 |
41 |
43 |
47 |
49 |
47 |
85 |
85 |
42 |
45 |
51 |
48 |
50 |
53 |
54 |
55 |
54 |
|
|
Wrch2 |
34 |
33 |
41 |
31 |
28 |
31 |
39 |
36 |
40 |
40 |
42 |
59 |
51 |
48 |
53 |
46 |
51 |
52 |
53 |
|
|
Wrch1 |
32 |
32 |
37 |
32 |
31 |
32 |
36 |
37 |
44 |
44 |
45 |
59 |
50 |
46 |
56 |
48 |
54 |
54 |
54 |
|
|
TC10 |
38 |
39 |
40 |
36 |
36 |
39 |
42 |
46 |
51 |
50 |
51 |
51 |
50 |
76 |
66 |
54 |
60 |
62 |
61 |
|
|
TCL |
35 |
37 |
39 |
34 |
35 |
35 |
38 |
44 |
48 |
49 |
48 |
48 |
46 |
76 |
63 |
53 |
58 |
60 |
59 |
|
|
Cdc42 |
40 |
40 |
42 |
37 |
37 |
38 |
43 |
43 |
53 |
51 |
50 |
53 |
56 |
66 |
63 |
61 |
69 |
71 |
70 |
|
|
RhoG |
39 |
40 |
40 |
37 |
41 |
41 |
44 |
46 |
55 |
55 |
53 |
46 |
48 |
54 |
53 |
60 |
72 |
72 |
70 |
|
|
Rac2 |
41 |
42 |
40 |
39 |
40 |
39 |
46 |
50 |
53 |
53 |
54 |
50 |
54 |
60 |
58 |
69 |
72 |
92 |
89 |
|
|
Rac1 |
42 |
42 |
41 |
39 |
41 |
42 |
49 |
49 |
57 |
57 |
55 |
52 |
54 |
62 |
60 |
71 |
72 |
92 |
93 |
|
|
Rac3 |
41 |
41 |
40 |
38 |
39 |
40 |
49 |
47 |
55 |
54 |
54 |
53 |
54 |
61 |
59 |
70 |
70 |
89 |
93 |
|
Group |
Rho Protein |
C-Terminal Sequence |
Lipid Modification |
Ref |
|---|---|---|---|---|
|
Typical |
RhoA |
KDGVREVFEMATRAALQARRGKKKSGCLVL |
GG |
[11] |
|
RhoB |
VREVFETATRAALQKRYGSQNGCINCCKVL |
GG, F, P |
||
|
RhoC |
KEGVREVFEMATRAGLQVRKNKRRRGCPIL |
GG |
||
|
Rac1 |
RGLKTVFDEAIRAVLCPPPVKKRKRKCLLL |
GG, P |
||
|
Rac2 |
RGLKTVFDEAIRAVLCPQPTRQQKRACSLL |
GG |
||
|
Rac3 |
RGLKTVFDEAIRAVLCPPPVKKPGKKCTVF |
GG |
||
|
RhoG |
QDGVKEVFAEAVRAVLNPTPIKRGRSCILL |
GG |
||
|
Cdc42 |
QKGLKNVFDEAILAALEPPEPKKSRRCVLL |
GG |
||
|
TCL |
AVFDEAILTIFHPKKKKKRCSEGHSCCSII |
F |
||
|
TC10 |
DEAIIAILTPKKHTVKKRIGSRCINCCLIT |
F, P |
||
|
Atypical |
RhoU |
QQQPKKSKSRTPDKMKNLSKSWWKKYCCFV |
P |
[11] |
|
RhoV |
EHKARLEKKLNAKGVRTLSRCRWKKFFCFV |
P |
||
|
RhoD |
AVFQEAAEVALSSRGRNFWRRITQGFCVVT |
F, GG |
[12] |
|
|
RhoF |
EDVFREAAKVALSALKKAQRQKKRRLCLLL |
F, GG |
||
|
Rnd1/ RhoS |
LSKRLLHLPSRSELISSTFKKEKAKSCSIM |
F |
[11] |
|
|
Rnd2/ RhoN |
MQRSAQLSGRPDRGNEGEIHKDRAKSCNLM |
F |
||
|
Rnd3/ RhoE |
KRISHMPSRPELSAVATDLRKDKAKSCTVM |
F |
||
|
RhoH/ TTF |
VFECAVRTAVNQARRRNRRRLFSINECKIF |
GG, F |
||
|
RhoBTB1 |
KREREKEDIALNKHRSRRKWCFWNSSPAVA |
Unknown |
N/A |
|
|
RhoBTB2 |
KRRWLFWNSPSSPSSSAASSSSPSSSSAVV |
Unknown |
|
Amino Acids 12, 59 and 61 |
|||
|---|---|---|---|
|
Group |
Subfamily |
Member |
Sequence |
|
Classic |
Cdc42 |
Cdc42 |
12 59 61 |
|
GDGAV---AGQED |
|||
|
Fast-cycling |
RhoU/RhoV |
RhoU |
GDGAV---AGQED |
|
RhoV |
GDGAV---AGQDE |
||
|
RhoD/RhoF |
RhoD |
GDGGC---AGQDD |
|
|
RhoF |
GDGGC---AGQED |
||
|
GTPase defective |
RhoBTB |
RhoBTB−1 |
GDNAV---FGDHH |
|
RhoBTB−2 |
GDNAV---FGDHH |
||
|
Rnd |
Rnd1 |
GDVQC---SGSPY |
|
|
Rnd2 |
GDAEC---SGSSY |
||
|
Rnd3 |
GDVQC---SGSPY |
||
|
RhoH |
GDSAV---AGNDA |
||
|
Amino acids 28 |
|||
|
Classic |
Rac |
Rac1 |
28 |
|
SYTTNAFPGEYIP |
|||
|
Fast-cycling |
RhoU/RhoV |
RhoU |
SYTTNGYPTEYIP |
|
RhoV |
SYTCNGYPARYRP |
||
|
RhoD/RhoF |
RhoD |
VFADGAFPESYTP |
|
|
RhoF |
VYSQGSFPEHYAP |
||
2. RhoH, An Atypical Rho Family Small GTPase
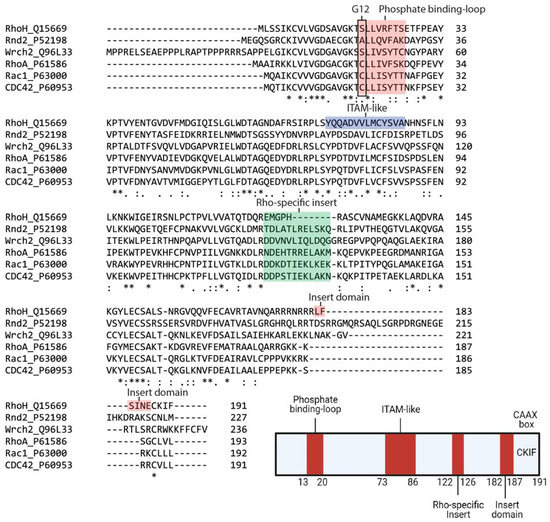
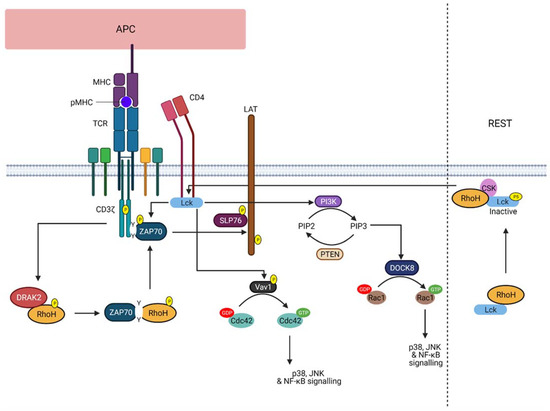
3. RhoH as a Therapeutic Target
References
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510.
- Fransson, Å.; Ruusala, A.; Aspenström, P. Atypical Rho GTPases Have Roles in Mitochondrial Homeostasis and Apoptosis. J. Biol. Chem. 2003, 278, 6495–6502.
- Boureux, A.; Vignal, E.; Faure, S.; Fort, P. Evolution of the Rho Family of Ras-Like GTPases in Eukaryotes. Mol. Biol. Evol. 2006, 24, 203–216.
- Aspenström, P.; Ruusala, A.; Pacholsky, D. Taking Rho GTPases to the next level: The cellular functions of atypical Rho GTPases. Exp. Cell Res. 2007, 313, 3673–3679.
- Valencia, A.; Chardin, P.; Wittinghofer, A.; Sander, C. The ras protein family: Evolutionary tree and role of conserved amino acids. Biochemistry 1991, 30, 4637–4648.
- Wennerberg, K.; Der, C.J. Rho-family GTPases: It’s not only Rac and Rho (and I like it). J. Cell Sci. 2004, 117, 1301–1312.
- Citalán-Madrid, A.F.; García-Ponce, A.; Vargas-Robles, H.; Betanzos, A.; Schnoor, M. Small GTPases of the Ras superfamily regulate intestinal epithelial homeostasis and barrier function via common and unique mechanisms. Tissue Barriers 2013, 1, e26938.
- Michaelson, D.; Silletti, J.; Murphy, G.; D’Eustachio, P.; Rush, M.; Philips, M.R. Differential localization of Rho GTPases in live cells: Regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 2001, 152, 111–126.
- Choy, E.; Chiu, V.K.; Silletti, J.; Feoktistov, M.; Morimoto, T.; Michaelson, D.; Philips, M.R. Endomembrane Trafficking of Ras: The CAAX Motif Targets Proteins to the ER and Golgi. Cell 1999, 98, 69–80.
- Adamson, P.; Paterson, H.F.; Hall, A. Intracellular localization of the P21rho proteins. J. Cell Biol. 1992, 119, 617–627.
- Ridley, A.J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006, 16, 522–529.
- Roberts, P.J.; Mitin, N.; Keller, P.J.; Chenette, E.J.; Madigan, J.P.; Currin, R.O.; Der, C.J. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 2008, 283, 25150–25163.
- Aspenström, P. Fast-cycling Rho GTPases. Small Gtpases 2020, 11, 248–255.
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9.
- Jaiswal, M.; Fansa, E.K.; Dvorský, R.; Ahmadian, M.R.; Communication, S.; Jaiswal, M.; Fansa, E.K.; Dvorsky, R.; Ahmadian, M.R. New insight into the molecular switch mechanism of human Rho family proteins: Shifting a paradigm. Biol. Chem. 2012, 394, 89–95.
- Shutes, A.; Berzat, A.C.; Cox, A.D.; Der, C.J. Atypical Mechanism of Regulation of the Wrch-1 Rho Family Small GTPase. Curr. Biol. 2004, 14, 2052–2056.
- Shutes, A.; Berzat, A.C.; Chenette, E.J.; Cox, A.D.; Der, C.J.B.T.-M. Biochemical Analyses of the Wrch Atypical Rho Family GTPases. Regul. Eff. Small Gtpases Rho Fam. 2006, 406, 11–26.
- Lin, R.; Bagrodia, S.; Cerione, R.; Manor, D. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 1997, 7, 794–797.
- Aspenström, P. Activated Rho GTPases in Cancer-The Beginning of a New Paradigm. Int. J. Mol. Sci. 2018, 19, 3949.
- Prive, G.G.; Milburn, M.V.; Tong, L.; de Vos, A.M.; Yamaizumi, Z.; Nishimura, S.; Kim, S.H. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc. Natl. Acad. Sci. USA 1992, 89, 3649–3653.
- Muñoz-Maldonado, C.; Zimmer, Y.; Medová, M. A Comparative Analysis of Individual RAS Mutations in Cancer Biology. Front. Oncol. 2019, 9, 1088.
- Fueller, F.; Kubatzky, K.F. The small GTPase RhoH is an atypical regulator of haematopoietic cells. Cell Commun. Signal. 2008, 6, 6.
- Chae, H.-D.; Siefring, J.E.; Hildeman, D.A.; Gu, Y.; Williams, D.A. RhoH regulates subcellular localization of ZAP-70 and Lck in T cell receptor signaling. PLoS ONE 2010, 5, e13970.
- Tamehiro, N.; Oda, H.; Shirai, M.; Suzuki, H. Overexpression of RhoH Permits to Bypass the Pre-TCR Checkpoint. PLoS ONE 2015, 10, e0131047.
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413.
- Troeger, A.; Chae, H.-D.; Senturk, M.; Wood, J.; Williams, D.A. A Unique Carboxyl-terminal Insert Domain in the Hematopoietic-specific, GTPase-deficient Rho GTPase RhoH Regulates Post-translational Processing. J. Biol. Chem. 2013, 288, 36451–36462.
- Li, X.; Bu, X.; Lu, B.; Avraham, H.; Flavell, R.A.; Lim, B. The Hematopoiesis-Specific GTP-Binding Protein RhoH Is GTPase Deficient and Modulates Activities of Other Rho GTPases by an Inhibitory Function. Mol. Cell. Biol. 2002, 22, 1158–1171.
- Horiguchi, H.; Ciuculescu, M.F.; Troeger, A.; Xu, H.; Brendel, C.; Williams, D.A. Deletion of Murine Rhoh induces More Aggressive Diffuse Large B Cell Lymphoma (DLBCL) Via Interaction with Kaiso and Regulation of BCL-6 Expression. Blood 2018, 132, 1574.
- Gu, Y.; Chae, H.-D.; Siefring, J.E.; Jasti, A.C.; Hildeman, D.A.; Williams, D.A. RhoH GTPase recruits and activates Zap70 required for T cell receptor signaling and thymocyte development. Nat. Immunol. 2006, 7, 1182.
- Muller, P.A.J.; Vousden, K.H.; Norman, J.C. p53 and its mutants in tumor cell migration and invasion. J. Cell Biol. 2011, 192, 209–218.
- Tajadura-Ortega, V.; Garg, R.; Allen, R.; Owczarek, C.; Bright, M.D.; Kean, S.; Mohd-Noor, A.; Grigoriadis, A.; Elston, T.C.; Hahn, K.M.; et al. An RNAi screen of Rho signalling networks identifies RhoH as a regulator of Rac1 in prostate cancer cell migration. Bmc Biol. 2018, 16, 1–20.
- Dorn, T.; Kuhn, U.; Bungartz, G.; Stiller, S.; Bauer, M.; Ellwart, J.; Peters, T.; Scharffetter-Kochanek, K.; Semmrich, M.; Laschinger, M.; et al. RhoH is important for positive thymocyte selection and T-cell receptor signaling. Blood 2007, 109, 2346–2355.
- Sanchez-Aguilera, A.; Rattmann, I.; Drew, D.Z.; Müller, L.U.W.; Summey, V.; Lucas, D.M.; Byrd, J.C.; Croce, C.M.; Gu, Y.; Cancelas, J.A.; et al. Involvement of RhoH GTPase in the development of B-cell chronic lymphocytic leukemia. Leukemia 2010, 24, 97–104.
- Tybulewicz, V.L.J.; Henderson, R.B. Rho family GTPases and their regulators in lymphocytes. Nat. Rev. Immunol. 2009, 9, 630–644.
- Kay, L. Characterisation of Atypical Human GTPases: Elucidation of Molecular Functions and Interactors. Ph.D. Thesis, Northumbria University, Newcastle, UK, 2016.
- Yablonski, D. Bridging the Gap: Modulatory Roles of the Grb2-Family Adaptor, Gads, in Cellular and Allergic Immune Responses. Front. Immunol. 2019, 10, 1704.
- Wang, H.; Zeng, X.; Fan, Z.; Lim, B. RhoH modulates pre-TCR and TCR signalling by regulating LCK. Cell. Signal. 2011, 23, 249–258.
- Pollyea, D.; Gore, L.; Gutman, J.; Eckhardt, S.G.; Hagelstrom, N.; Coutre, S.; Thirman, M.; Byrd, J. A Dose Escalation Study of Ibrutinib with Lenalidomide for Relapsed and Refractory Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma. Ann. Oncol. 2013, 24, i33.
- Troeger, A.; Johnson, A.J.; Wood, J.; Blum, W.G.; Andritsos, L.A.; Byrd, J.C.; Williams, D.A. RhoH is critical for cell-microenvironment interactions in chronic lymphocytic leukemia in mice and humans. Blood 2012, 119, 4708–4718.
- Robert, G.; Jacquel, A.; Auberger, P. Chaperone-Mediated Autophagy and Its Emerging Role in Hematological Malignancies. Cells 2019, 8, 1260.
- Anguiano, J.; Garner, T.P.; Mahalingam, M.; Das, B.C.; Gavathiotis, E.; Cuervo, A.M. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 2013, 9, 374–382.
- Wagner, M.; Oelsner, M.; Moore, A.; Götte, F.; Kuhn, P.-H.; Haferlach, T.; Fiegl, M.; Bogner, C.; Baxter, E.J.; Peschel, C.; et al. Integration of innate into adaptive immune responses in ZAP-70–positive chronic lymphocytic leukemia. Blood 2016, 127, 436–448.
- Burger, J.A.; Chiorazzi, N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013, 34, 592–601.
- Kohlhaas, V.; Blakemore, S.J.; Al-Maarri, M.; Nickel, N.; Pal, M.; Roth, A.; Hövelmeyer, N.; Schäfer, S.C.; Knittel, G.; Lohneis, P.; et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood 2021, 137, 646–660.
- Visperas, P.R.; Wilson, C.G.; Winger, J.A.; Yan, Q.; Lin, K.; Arkin, M.R.; Weiss, A.; Kuriyan, J. Identification of Inhibitors of the Association of ZAP-70 with the T Cell Receptor by High-Throughput Screen. SLAS Discov. Adv. Life Sci. R D 2017, 22, 324–331.
- Boohaker, R.J.; Lee, M.W.; Vishnubhotla, P.; Perez, J.M.; Khaled, A.R. The use of therapeutic peptides to target and to kill cancer cells. Curr. Med. Chem. 2012, 19, 3794–3804.
- Cooper, B.M.; Iegre, J.; O’ Donovan, D.H.; Ölwegård Halvarsson, M.; Spring, D.R. Peptides as a platform for targeted therapeutics for cancer: Peptide–drug conjugates (PDCs). Chem. Soc. Rev. 2021, 50, 1480–1494.